. Introduction
Common hazel (Corylus avellana L., Betulaceae) is an economically important nut crop in many countries in the world owing to the nutritional and nutraceutical properties of its nuts (Arzanlou et al., 2018). The plant occurs in the wild throughout Europe, South America, and Asia Minor to the Caucasus. In Poland, it is common both in the lowlands and in the mountains up to about 1,300 m above sea level (Rolnik & Olas, 2018; Zając & Zając, 2001). Due to its low soil requirements, it grows in natural stands in deciduous and mixed forests as well as coniferous forests (Rolnik & Olas, 2018). Hazelnuts are one of the most popular nuts cultivated in the world. Poland is one of the largest producers of hazelnuts in Europe, with production of approx. 5,300 tonnes per year and a yield of 1.4 t ha−1 (Król et al., 2020).
Hazel trees are attacked by various pathogens. In Europe, Pseudomonas avellana and Pseudomonas syringae pv. coryli are responsible for bacterial canker and hazelnut dieback. In addition, in some European countries, such as Spain and Poland, C. avellana is affected by apple mosaic virus (Enescu et al., 2016).
Many hazel diseases are caused by fungal pathogens. Amongst one of the most numerous causal agents are powdery mildew fungi. Erysiphe corylicola U. Braun and S. Takam., E. verruculosa (Y.N. Yu & Y.Q. Lai) U. Braun & S. Takam., E. ellisii (U. Braun) U. Braun and S. Takam. belonging to Erysiphe sect. Microsphaera can occur on the representatives of the genus Corylus (Braun & Cook, 2012). Based mainly on phylogenetic analyses, Bradshaw et al. (2021) introduced two new Erysiphe species infesting Corylus plants, i.e., E. cornutae M. Bradshaw (separated from E. corylacearum as a new species) and E. coryli-americanae M. Bradshaw. However, the most common species of powdery mildew fungus recorded on hazel in Poland and worldwide is Phyllactinia guttata(Wallr.: Fr) Lév. In the course of the research on P. guttata, other symptoms were accidentally discovered on hazel leaves, and after careful identification, the parasite was found to be a new species for Poland. P. guttata usually develops its mycelium on the lower side of the leaves, while E. corylacearum forms mycelium also on the upper side (amphigenous). So far, P. guttata has been the main cause of disease in hazel in Poland. For some time, powdery mildew species regarded as new to Poland have been found sporadically and have usually been associated with alien species migration, mostly from Asia.
. Material and methods
Sample collection and morphological identification
Plant leaves affected by E. corylacearum were collected in 2018, 2022, and 2023 in Lublin (Poland). The material was air dried, and the morphological structures of the specimens were examined in microscopic preparations stained with Lactophenol Cotton Blue dye and observed using an Olympus BX53 light microscope with a camera attachment. Measurements of ca. 50 chasmothecia, appendages, asci, ascospores, and conidia were made, and the structures were photographed using a microscope Olympus digital camera SC180 and a scanning electron microscope (SEM) TESCAN vega 3 LMU (Brno, Czech Republic). The collected specimens were deposited in the herbarium of Maria Curie-Skłodowska University in Lublin (LBL). The results of morphological analysis were compared with the species descriptions in Braun (1987) and Braun and Cook (2012). Samples containing the morphological structures of the fungus (mycelium, chasmothecia with asci and ascospores, conidiophores, and conidia) were then prepared for further genetic analyses under the control of an Olympus SZ61 stereoscopic microscope.
Genetic analysis
DNA isolation
DNA isolation from fungal cells was carried out using a commercial DNeasy Plant Mini Kit (Qiagen) in accordance with the manufacturer’s instruction. The purity and concentration of genomic DNA were checked by NanoDropTM 2000/2000c measurements (Thermo Fisher Scientific, USA). The extracted DNA was stored at −20 °C.
Molecular phylogeny
The phylogenetic analysis of the fungal isolate was determined by comparative analysis of partial sequences of the ITS (internal transcribed spacer) region and LSU (large subunit) sequences of the rRNA gene. These sequences were amplified using primer pairs ITS1/ITS4 and PM3/TW14 (Hsiao et al., 2022; Takamatsu & Kano, 2001; White et al., 1990). All analyzed sequences were amplified in the PCR reaction using a ReadyMixTM Taq PCR Reaction Mix kit (Sigma-Aldrich) following the manufacturer’s instruction. 50 ng of template DNA and 0.4 mM of forward and reverse primers were added to the reaction mixture. PCR amplifications were performed using a thermal cycler (Perkin Elmer Cetus, Norwalk, USA) in the following conditions: initial denaturation at 94 °C for 5 min, followed by 35 cycles of denaturation at 94°C for 45 s (ITS)/30 s (LSU), annealing at 46 °C for 45 s (ITS)/55 °C for 1 min (LSU), followed by an extension step of 1 min 40 s at 72 °C (ITS)/1 min at 72 °C (LSU), with a final extension step at 72 °C for 8 min.
The amplicons were purified on columns using a Clean-Up purification kit (A&A Biotechnology) and sequenced with a Terminator Cycle sequencing kit. The sequencing reactions were read using a 3500 Genetic Analyzer according to the procedure specified by the producer (Life Technologies). The sequences were compared with relevant sequences in the GenBank database using the BLAST tool. The ClustalX2 program was used for sequence alignments analyzed in the GeneDoc program (Nicholas et al., 1997; Thompson et al., 1994). A phylogenetic tree was constructed based on the generated sequence set using the MEGA11 program with the Neighbor-Joining method (Tamura et al., 2021). The two-parameter Kimura model was used as a nucleotide substitution model. The jModelTest selected the best-fitting evolutionary model for each tested gene (Darriba et al., 2012). The statistical significance of the tree was evaluated with the bootstrap test (1,000 replicates). The phylogenetic tree was represented in the TreeView program (Page, 1996).
. Results and discussion
Morphological characteristics
Erysiphe corylacearum U. Braun and S. Takam., in Braun, Schlechtendalia 8:33 (2002) (Figure 1).
Figure 1
Erysiphe corylacearum on Corylus avellana. (A) Infected leaves in vivo; (B) Conidia; (C) Conidium in SEM; (D) Chasmothecia in SEM; (E) Chasmothecium; (F) Asci with ascospores. (B, E–F) Structure in a microscopic preparation stained with cotton blue in lactic acid. Scale bars: (A) 10 mm; (B, F) 20 µm; (C) 10 µm; (D) 100 µm; (E) 50 µm. Herbarium collections: (A, B–C) LBL M–33132; (D–F) LBL M–33130. Photographs: U. Świderska.

Mycelium amphigenous, whitish, forming dense patches. Conidiophores with dimensions of 64–80 × 7–9 µm. Solitary conidia formed on conidiophores, ellipsoid to ovoid, hyaline, 25–32 × 16–20 µm in size. Chasmothecia gregarious to scattered on both sides of leaves, 79–90(–100) µm in diameter; appendages usually up to 11, aseptate or with only single septum at the base, 3–5 times branched on the top, mostly of the same length as the chasmothecial diameter. Asci obovoid to broadly ellipsoid, usually four with 7–8-spores, with dimensions of 43–49 × 36–40 µm. Ascospores ellipsoid or ovoid, hyaline, with dimensions of (16–)19–25 × 11–14.5 µm.
Specimens examined: On Corylus avellana L. (Betulaceae). POLAND. Lublin Voivodeship: Lublin – Głęboka St., 6 Aug. 2018, leg. A. Wołczańska (LBL M–33131), 30 Aug. 2022, leg. U. Świderska (LBL M–33130), 23 June 2023, leg. U. Świderska (LBL M–33132).
GenBank sequence accession numbers: The GenBank accession numbers for the sequences reported are LSU: OR272003 and ITS: OR272002 (for herbarium collection number LBL M–33130). The accession numbers of the reference strains are given on the phylograms.
C. avellana is a common species occurring naturally in forests and grown as an ornamental and usable plant in Poland. The occurrence of powdery mildew on their leaves has long been observed, with P. guttata found most frequently to date. In 2018, some mycelium was observed on the upper side of the leaves, unlike in the case of P. guttata parasitism; therefore, the infected leaves were collected, and the fungus was identified. Preliminary microscopic identification excluded belonging to the genus Phyllactinia, which is characterized by, among others, straight, rigid, acicular appendages with characteristic bulbous swelling at the base (Braun & Cook, 2012). Although other fungal species of the genus Erysiphe may occur on representatives of the genus Corylus, only E. corylacearum has so far been recorded on C.avellana worldwide and currently in Poland. The morphological characters of the examined sample correspond to the description of E. corylacearum in Braun and Cook (2012) and Bradshaw et al. (2021). These features also agree with the descriptions of the species published, especially from Europe since 2019 (e.g. Beenken et al., 2020; Boneva et al., 2023; Kalmar et al., 2022; Mezzalama et al., 2021; Rosati et al., 2021; Voglmayr et al., 2020; Zajc et al., 2023). The morphological and molecular analyses of the collected material confirmed the presence of the new species in Poland.
The disease caused by E. corylacearum was first noted in Asia (China, South Korea, Japan, Turkey, Iran, Georgia, Azerbaijan, Ukraine, North Korea, and Russia) and since 2019 in Europe (Zajc et al., 2023). In terms of the timeline of its occurrence in Europe, the fungus was successively reported in Ukraine and Russia (Heluta et al., 2019), Austria (Voglmayr et al., 2020), Switzerland (Beenken et al., 2020), Italy (Mezzalama et al., 2021), Romania (Rosati et al., 2021), Hungary (Kalmar et al., 2022), Slovenia (Zajc et al., 2023), and Bulgaria (Boneva et al., 2023).
In order to determine the taxonomic position and phylogenetic relationships of the C. avellana isolate, initially designated as a representative of the powdery mildew family, with other members of this family, the sequences of the LSU and ITS regions of the isolate were compared to the corresponding sequences of reference strains available in the Genbank database.
The study based on LSU sequence analysis included 35 representatives of the Erysiphaceae family. The analysis compared the sequences of the LSU regions of the tested isolate with the corresponding sequences available in the GenBank database, and the phylogenetic tree was built using sequencing data of the LSU region. The analyzed strain, E. corylacearum, was grouped with strains representing the genus Erysiphe (with 100% bootstrap support) (Figure 2). The sequence similarity of the isolate from C. avellana to the sequence of reference strains representing the genus Erysiphe ranged from 90 to 100%.
Figure 2
Phylogenetic relationships of partial LSU gene sequences determined by Neighbor-Joining gene phylogeny. Numbers at the nodes indicate the levels of bootstrap support based on Neighbor-Joining analysis of 1,000 resampled datasets. Bootstrap values ≥50% are given at the branching points. Reference sequences were obtained from the GenBank database (accession numbers are given in parentheses).
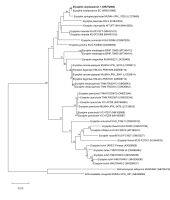
In the phylogenetic tree, all strains of the genus Erysiphe formed a cluster with 100% bootstrap support. The E. corylacearum strain isolated from C. avellana, i.e., a plant originating from the eastern part of Poland (Lublin), formed (with bootstrap support of 94%) a cluster with the E. corylecearum EC strain isolated from a C. avellana plant originating from Austria (Figure 2). The LSU sequence similarities of the isolate and other members of the family Erysiphaceae, i.e., Arthrocladiella and Golovinomyces, were in the range of 89 and 82%, respectively.
The study involved 44 representatives of the Erysiphaceae family. The taxa included 20 species representing three genera isolated from various plant species. The phylogenetic ITS sequence analysis has confirmed that the isolate from C. avellana belongs to the family of mildew fungi (Erysiphaceae) and represents the genus Erysiphe. It showed 91–99% similarity of the ITS sequence to other representatives of this genus. On the phylogenetic tree (Figure 3), it formed a common cluster with strains representing different Erysiphe species with bootstrap support of 100%. In the phylogenetic trees, the isolate formed a well-supported monophyletic clade (99% bootstrap support) with E. corylacearum strains (Figure 3). A separate group on the phylogenetic tree was represented by fungi of the genera Golovinomyces and Arthrocladiella. The degree of similarity of the nucleotide sequence of the isolate’s ITS sequence to the sequences of representatives of the genera Golovinomyces and Arthrocladiella was 71% and 75%, respectively. These results confirm that the isolate belongs to the genus Erysiphe and simultaneously indicate that this isolate is a member of the E. corylacearum species.
Figure 3
Phylogenetic relationships of partial ITS sequences determined by Neighbor-Joining gene phylogeny. Numbers at the nodes indicate the levels of bootstrap support based on Neighbor-Joining analysis of 1,000 resampled datasets. Bootstrap values ≥50% are given at the branching points. Reference sequences were obtained from the GenBank database (accession numbers are given in parentheses).

E. corylacearum has already been recorded as a serious invasive alien pathogen, causing significant losses in hazelnut crops in Asia (i.e., Turkey, Iran, and Georgia) (Arzanlou et al., 2018; Voglmayr et al., 2020). Subsequent reports of the occurrence of this species of fungus in Europe prove that it is an increasingly spreading species and a threat to naturally occurring and especially cultivated hazelnuts also in Poland. Further records of the fungus are to be expected, and it should be observed whether it will oust the most common species P. guttata.


