Introduction
The genus Hodophilus R. Heim ex R. Heim was recently recovered by Birkebak et al. (2016) based on the polyphyly of agaricoid members of the family Clavariaceae. It can be distinguished from Camarophyllopsis Herink s. str. and Lamelloclavaria Birkebak & Adamčík by a hymeniderm pileipellis composed of broadly inflated (globose, obpyriform to sphaero-pendunculate) terminal elements, with predominantly parietal pigment and the absence of clamps (Arnolds, 1986, 1990; Birkebak et al., 2016; Horak, 1968). The genus Hodophilus, hitherto unknown in Poland, is known to comprise approximately 26 species worldwide (Adamčík, Jančovičová, Looney, Adamčíková, Birkebak, et al., 2017; Adamčík, Jančovičová, Looney, Adamčíková, Griffith, et al., 2017; Adamčík et al., 2016, 2018, 2020; Crous et al., 2017), 15 of which are in Europe (Adamčík et al., 2018, 2020; Adamčík, Jančovičová, Looney, Adamčíková, Birkebak, et al., 2017; Adamčík, Jančovičová, Looney, Adamčíková, Griffith, et al., 2017).
During field mycological studies held in 2014 and subsequently in 2016 in a transitional area of vegetation between rupicolous calcareous grasslands (Origano-Brachypodietum laserpitietosum, Origano-Brachypodietum stachyetosum) and thermophilous beech forest (Carici albae-Fagetum) in the Pieniny Mts, the first author found pale brown to grayish brown agaricoid fruiting bodies of an omphalioid habit. Laboratory studies (based on microscopy and sequencing) have shown that they are probably one of the most common species of the genus Hodophilus, that is, Hodophilus variabilipes Jančovičová, Adamčík & Looney, a fungus recently described from material from widely scattered localities in several European countries, including the Czech Republic, Denmark, France, the United Kingdom, Slovakia (Malé Karpaty Mts; holotypus), and Sweden (Adamčík, Jančovičová, Looney, Adamčíková, Griffith, et al., 2017).
The primary goals of the present study were to investigate and describe the first collections of H. variabilipes from Poland using morphological and molecular characteristics. We used phylogenetic analysis of nuclear ribosomal DNA (ITS) sequences to determine the phylogenetic position of the study material. We also aimed to provide new insight into the European distribution and ecology of this species.
Material and Methods
Morphological Studies
Field photographs of fresh basidiomata were taken with the aid of a Panasonic Lumix DMC-FZ50 camera equipped with a Raynox DCR-250 Super Macro. Characteristics that change over time (color, smell, and texture of basidiomata) were noted in the field. The macroscopic description is based on both the study of fresh material and the analysis of photos. Microcharacters were observed with a light microscope equipped with a digital camera. All microscopic structures were observed in the dried material. Freehand sections of rehydrated basidiomata pieces were examined in squash preparations in 5% NH3·H2O, 5% KOH, 1% Phloxine B in 5% NH3·H2O, and Melzer’s reagent. Image-grabbing and biometric analyses were performed using the NIS-Elements D 3.1 imaging software. The dimensions of the microcharacters are presented as follows: (minimum) 10–90 percentile values (maximum), average ± standard deviation. For basidiospores, randomly selected mature spores were measured without the hilar appendix. The length of the basidia was measured excluding the sterigmata. The following abbreviations are used: L = number of lamellae reaching the stipe, l = number of lamellulae between each pair of lamellae, and Q = the length-width ratio of basidiospores (mean value). Morphological terminology reported by Vellinga (Vellinga, 1988, 1990) and Arnolds (1990) was used. The nomenclature of vascular plants followed the checklist provided by Mirek et al. (2002) and that of plant communities followed the checklist provided by Pancer-Koteja et al. (2004). The nomenclature of fungi was based on the Index Fungorum (http://www.indexfungorum.org/). The studied collections were deposited in the Museum of Natural History, University of Wrocław, Poland (WRSL).
Molecular Procedures
DNA Extraction, PCR Amplification, and DNA Sequencing
The ITS rDNA region (containing ITS1-5.8S-ITS2 sequences), which is commonly used for the separation and identification of agaric species (e.g., Adamčík, Jančovičová, Looney, Adamčíková, Griffith, et al., 2017; Borovička et al., 2012; Miller & Buyck, 2002; Schoch et al., 2012; Zhang et al., 2004), was used to confirm the taxonomic affiliation of the collected material and to infer the evolutionary relationships between H. variablipes and other representative taxa in this study. Genomic DNA isolates were extracted from two dried specimens (ChP-2014-0007, ChP-2016-0004) using the CTAB procedure described by Murray and Thompson (1980). Spectrometric DNA concentration measurements were conducted using a NanoDrop ND 2000C (Thermo Scientific). Then, DNA isolates were purified with a Syngen DNA clean-up Kit (Syngen Biotech) to obtain good quality templates, and the DNA concentration was measured again.
ITS rDNA regions were amplified using the primers ITS1F and ITS4 or ITS1 and ITS4-BR (Gardes & Bruns, 1993; White et al., 1990). PCR was performed in a 20-μL volume with Q5 High-Fidelity DNA Polymerase (New England BioLabs) according to the manufacturer’s protocol.
Sanger sequencing was carried out with the BrilliantDye Terminator v3.1 Kit (NimaGen). Reactions were set with fourfold dilution of the reaction premix and addition of BrilliantDye Terminator 5X Sequencing Buffer (NimaGen) according to the manufacturer’s instructions. Sequencing products were separated by capillary electrophoresis on an Applied Biosystems 310 Genetic Analyzer (Thermo Fisher Scientific). Two sequences were generated for this molecular and phylogenetic analysis, along with additional sequences obtained from NCBI GenBank (https://www.ncbi.nlm.nih.gov/).
Sequence Alignment and Phylogenetic Analysis
Sequence Scanner Software ver. 2.0 was used for basecalling, and peak quality was also checked using the Sanger Quality Check App (Thermo Fisher Scientific). Sequences were manually assembled in BioEdit Sequence Alignment Editor (Hall, 1999) and deposited in GenBank. All obtained sequences were compared with sequences stored in the GenBank database using the BLAST algorithm (Altschul, 1990). Based on the BLASTN results, sequences from foreign collections were selected according to the outcomes of recent phylogenetic studies on the most closely related members of Hodophilus (Adamčík, Jančovičová, Looney, Adamčíková, Griffith, et al. 2017). ITSx (Bengtsson-Palme, 2013), as implemented on the PlutoF web-based workbench (Abarenkov et al., 2010), was then used to cut off flanking gene fragments from all sequences assembled in a data matrix. Prior to the phylogenetic analyses, the ITS1-5.8S-ITS2 DNA sequences were aligned using MAFFT (Katoh et al., 2002, 2005) on MyHits gateway (http://myhits.isb-sib.ch/), with default conditions for gap openings and gap extension penalties and subsequently adjusted by eye in MEGA ver. 7.0 (Tamura et al., 2013). The alignment was then entered into the program Gblocks ver. 0.91b (Castresana, 2000; Talavera & Castresana, 2007) to objectively eliminate poorly aligned positions and divergent regions using the least stringent parameters. Likelihood (ML) and Bayesian inference (BI) optimality criteria were determined with RAxML ver. 8.2.10 (Stamatakis, 2006, 2014) operated via raxmlGUI ver. 1.5b (Silvestro & Michalak, 2012) and MrBayes ver. 3.2.2 (Ronquist et al., 2012), respectively. jModelTest 2 software (Posada, 2008) was used to identify the sequence evolution model that fit the data set using both the Akaike information criterion (AIC) and the Bayesian information criterion (BIC). Finally, the HKY+Γ model of nucleotide substitution was applied to all unlinked partitions (ITS1 spacer, 5.8S gene, ITS2 spacer) in the ML analysis, and GTR+Γ (ITS1), K80+I (5.8S), and HKY+Γ (ITS2) models were used in BI inference. Ramariopsis corniculata (KM248910) was chosen as the outgroup taxon for the analyzed dataset. For ML analysis to perform a tree inference and search for the optimal topology, 103 bootstrap replicates were set using the rapid bootstrap algorithm (Stamatakis, 2008). Support values from bootstrapping runs (MLB) were reported on the globally best tree. BI was performed with four (three incrementally heated and one cold) Monte Carlo Markov chains (MCMC) that were independently run twice, sampling one tree every 100 generations for 2×106 generations, starting with a random tree. Completion was determined by the average standard deviation of split frequencies falling below 0.003. The first 25% of generations were discarded as burn-in, whereas the remaining trees were used to calculate a 50% majority-rule tree and to determine the posterior probabilities (BPP) of individual branches. Significance thresholds were set above 50% for bootstrap and 0.90 for posterior probability. Consensus trees were visualized and compared using TreeGraph ver. 2.14b (Stöver & Müller, 2010). To analyze the intra- and interspecific genetic variability of Hodophilus species, uncorrected pairwise distances (p distance) were obtained using MEGA ver. 7.0 (Tamura et al., 2013).
Results
Morphological Description of the Material
Hodophilus variabilipes Jančovičová, Adamčík & Looney, Mycological Progress 16 (8): 818 (2017).
Illustrations: Adamčík, Jančovičová, Looney, Adamčíková, Griffith, et al. (2017, Figures 6–9, 16–22).
Pileus 6–20 mm, convex with deflexed or inflexed margin, then plano-convex to applanate, with straight margin, hygrophanous, when moist at center dark brown, reddish or grayish brown with paler – yellowish brown to grayish brown margin, on drying pale grey-brown to beige, not or indistinctly translucently striate up to one third way of the radius when wet, surface matt, velvety, or appearing granular under lens, at first smooth, then becoming rugose or rough towards the center, sometimes with concentrical cracks around the margin. Lamellae, L = 15–24, 1 = 1–3, distant, thick, arcuate, short to deeply decurrent, sometimes furcate, beige, light brown when young, when mature rather dark grayish brown, edge entire, concolorous, or slightly paler than the sides. Stipe 15 × 25 mm, cylindrical or tapering towards base, sometimes with swollen apex, more or less flexuous, stuffed or narrowly fistulose, usually concolorous along the entire length (sometimes whitish tomentose at the base), grayish yellow, yellowish brown to dark brown, with fine brown to dark brown squamules along the entire length, contrasting with paler background, rarely smooth. Context firm, elastic, concolorous with the surface, pale beige to light brown inside. Smell and taste are not distinctive.
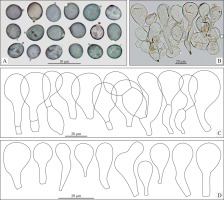
Basidiospores (4.4)4.8–5.7(5.9), 5.2 ± 0.4 × (3.8)4.2–4.8(5.4), 4.5 ± 0.3 µm, Q = (1)1–1.3(1.4), 1.2 ± 0.1 (n = 60 spores per two collections), in the majority broadly ellipsoid, some subglobose or ellipsoid, with small but distinct, rather blunt hilar appendage, hyaline, smooth, thin-walled, inamyloid, sometimes weakly to strongly dextrinoid. Basidia (27.9)31.7–43.5(44.3), 37.7 ± 4.7 × (4.5)4.8–7.1(7.2), 6.4 ± 0.9 µm (n = 17), narrowly to very narrowly clavate, with four sterigmata, usually flexuous towards the base. Marginal cells on the lamellar edges well or poorly differentiated: (16)17–31.5(38), 23 ± 6 × (6.5)8–12.5(16), 10 ± 2 µm (n = 32), usually broadly clavate or obpyriform, rarely almost sphaeropedunculate. Pleurocystidia were not found. Pileipellis near the pileus margin a transition from hymeniderm to the epithelium, pileipellis near the pileus center a hymeniderm, made up of erect hyphae with short, inflated elements, broader towards apex, with usually obpyriform or broadly clavate, rarely subglobose or ellipsoid terminal elements, (18.5)25–42(50) × (15)18.5–28.5(35), 23 ± 3.5 × (15)18.5–28.5(35), 23 ± 3.5 µm (n = 72), with yellowish brown parietal pigment. Caulocystidia are abundantly present, mainly broadly clavate, and have not been studied in detail. Stipitipellis a cutis of repent, cylindrical hyphae not studied in detail. Clamp-connections are absent.
Specimens examined: POLAND. 1. Pieniny Mts, Pieniny National Park, Sromowce Niżne village, in the Pieniny gorge, on the back of the rock Grabczychy (49°24′27.97″ N, 19°25′08.12″ E), 580 m a.s.l., a dozen of basidiomata on the ground, in the ecotone area between rupicolous calcareous meadow of Origano-Brachypodietum laserpitietosum and stachyetosum associations and thermophilous beech forest Carici albae-Fagetum (in Picea abies thickets with scattered Corylus avellana and Juniperus communis), September 6, 2014, leg. P. Chachuła (ChP-2014-0007; GenBank: MK434303). 2. Ibidem, August 24, 2016, leg. P. Chachuła (ChP-2016-0004; GenBank: MK434304).
Molecular Characterization
Sanger sequencing of the rDNA containing the internal transcribed spacer (ITS1, ITS2) regions and the 5.8S coding sequence of two H. variabilipes collections, ChP-2014-0007 and ChP-2016-004, yielded 506 and 513-bp-long fragments, respectively. The aligned ITS complete data set consisted of 35 sequences (one as the outgroup) representing 14 taxa and 643 characters. After excluding ambiguous (mainly terminal) ITS-regions of the data set, 483 characters remained for the final analysis. Of these, 304 were constant, 107 were parsimony-informative, and 72 were parsimony-uninformative. Both types of analysis, Bayesian inference (BI), and maximum likelihood (ML) revealed almost identical trees topology, and thus only the tree inferred from the BI strategy is shown in Figure 3. The phylogram shows that H. variabilipes forms a well-supported group (BPP = 0.95, MLB = 86) together with H. micaceus, H. subfuscescens, and H. albofloccipes. However, our data poorly resolved the relationships of the H. variabilipes and H. micaceus clade because of the lack of sufficient phylogenetic information within the ITS data. The sequences obtained in this study (MK434303, MK434304) are within the H. variabilipes clade together with other sequences of H. variabilipes, including the holotype sequence from Slovakia (GenBank NR158523). The estimated interspecific nucleotide differences of H. variabilipes inferred from all studied ITS sequences varied from 0.2% to 0.7%. The percentage of the ITS sequence difference between the Polish material and the material represented by vouchers from the type country (GenBank MF043549, MF043539, NR158523, MF043548, and MF043547) was 0.2%. A BLASTN search using the Polish ITS sequences (506 bp and 513 bp) of H. variabilipes showed H. variabilipes from Denmark (GenBank MF043546; 99% identity) as the closest hit.
Figure 3
Phylogenetic placement of Hodophilus variabilipes Jančovičová, Adamčík & Looney inferred from ITS rDNA data. The best tree resulting from Bayesian inference analysis is presented with BPP (Bayesian posterior probabilities values) ≥ 0.90 and MLB (maximum likelihood bootstrap percentage) ≥ 50 displayed on the branches. Sequences printed in bold were obtained during this study. The GenBank accession number for each sequence is given in brackets next to the species name. Also included are country names and whether the DNA sequence represents a type collection (labeled by asterisk). Ramariopsis corniculata was used as an outgroup.

Discussion
Until quite recently, H. atropunctus (Pers.: Fr.) Birkebak & Adamčík was defined as the only species of the genus with distinct dark colored dots on the stipe (Adamčík, Jančovičová, Looney, Adamčíková, Birkebak, et al., 2017; Bas et al., 1990; Boertmann, 2012; Bon, 1977; Hesler & Smith, 1963; Horak, 2005; Kovalenko et al., 2012; Printz & Læssøe, 1986). However, multilocus phylogenetic studies conducted by Adamčík, Jančovičová, Looney, Adamčíková, Griffith, et al. (2017) allowed the recognition of two distinct taxonomic entities morphologically corresponding to the traditionally defined H. atropunctus concept. Thus, H. atropunctus was assigned to the paler of the two entities, while the darker one was described as a new species, H. variabilipes. It appears that these two odorless species are best distinguished by molecular markers, but morphological traits unique for each species are also defined. According to Adamčík, Jančovičová, Looney, Adamčíková, Griffith, et al. (2017), the color of basidiomata and the color changes during maturity and the humidity of basidiomata are the key diagnostic characteristics for both species. Hodophilus variabilipes is characterized as having a darker brown pileus when young and fresh and a darker lamella color in mature conditions (brown to dark brown). The discoloration (fade) of the pileus in H. variabilipes starts near the center (not from the pileus margin), and the pileus is eventually uniformly discolored in dry conditions. Moreover, in contrast to the stipe of H. atropunctus, which usually has a distinctly paler color near the apex, the stipe of H. variabilipes displays a more or less uniform color pattern. Both species share rather similar microscopic structures, although H. variabilipes has less elongated spores (av. Q = 1.22). It is worth mentioning that specimens of H. variabilipes without darker dots on the stipe are sometimes found, and the species might hence be confused with other taxa of the H. micaceus superclade because of similar macromorphology (Adamčík, Jančovičová, Looney, Adamčíková, Griffith, et al., 2017).
The present study is the first to report a member of the genus Hodophilus in Poland. This may be surprising, as Kreisel (2011) reported four Hodophilus species from the neighboring region, Mecklenburg-Vorpommern. The morphology and molecular identification based on ITS rDNA sequences confirmed that the fungus collected in Poland belongs to H. variabilipes. In general, the micro- and macrocharacters of the studied specimens match the description and iconography of H. variabilipes given by Adamčík, Jančovičová, Looney, Adamčíková, Griffith, et al. (2017). However, the variation in the basidiospore reaction to Melzer’s reagent disagrees with the original concept that the species has no dextrinoid spores (Adamčík, Jančovičová, Looney, Adamčíková, Griffith, et al.,2017). It was surprising that, among the studied collections, the majority of specimens had a smaller or larger proportion of dextrinoid basidiospores, that is, those with reddish brown to rust brown color in Melzer’s reagent. This (diffuse) iodine reaction varied from weak to strong and was visible in a few to many spores. The appearance of this reaction type is unexpected, since this feature has not been reported in previous works on the Hodophilus, and not obvious. It is worth mentioning that such reactions, when a drop of Melzer’s iodine reagent is added, have also been noted in Dermoloma, in species with inamyloid spores (the section Dermoloma J. E. Lange, i.e., D. cunefifolium), where it was supposed that the occurrence of the dextrinoid reaction is associated with the ripening of spores (Arnolds, 1992). It is striking that the dextrinoid reaction was only observed in the specimens assigned to H. variabilipes, and it was not confirmed in the case of some collections tentatively identified as H. atropunctus (material not included in this study, and not verified molecularly). However, it is uncertain whether this variation is taxonomically useful, and further studies are required. The results of our molecular analyses agree with previous molecular data (Adamčík et al., 2018; Adamčík, Jančovičová, Looney, Adamčíková, Griffith, et al., 2017) that suggested close relationships between fungi of the H. micaceus clade and the H. variabilipes group. Contrary to the close relationship between the species groups, we also confirmed the large phylogenetic distance between H. variabilipes and H. atropunctus. Our studies highlight a low intraspecific genetic diversity within the H. variabilipes clade (0.2%–0.7%), and the same goes for the percentage of the ITS sequence difference between the holotype and the Polish material (0.2%). However, we stress that the inferences drawn from the provided ITS molecular analyses are only provisional, pending multigene analyses utilizing multiple loci, such as nLSU rDNA, rpb, and tef, which may support or change the conclusions reported above and may also allow for a better understanding of the noted discrepancies in results.
Despite the fact that until now, H. variabilipes has been known to originate only from six European countries, the distribution of the species might be significantly wider. It is likely, however, that Hodophilus species with darker dots on the stipe are limited to temperate areas of Europe or Eurasia, because they have not been reported in North America (Adamčík, Jančovičová, Looney, Adamčíková, Griffith, et al., 2017; Hesler & Smith, 1963). Hodophilus variabilipes and H. atropunctus may share common habitat types, and, according to Adamčík, Jančovičová, Looney, Adamčíková, Griffith, et al. (2017), their tendency to occur near edges between forested areas and meadows is noteworthy. In general, Hodophilus species prefer black and clay soils, often with a very high pH (Adamčík et al., 2018). Our study confirmed that this is true for H. variabilipes reported from the Pieniny Mts, where the species was found within the ecotone area between natural xerothermic grasslands and thermophilous beech forest, on rich soils developed over limestone rocks.
It is not easy to explain why fungi with such a wide distribution are scattered, rare, or even absent from large areas of Europe, where suitable habitats are not rare. Although both H. variabilipes and H. atropunctus may be overlooked in places because of a superficial likeness to a host of little brown mushrooms (LBMs), their distribution pattern is probably strongly influenced by specific microclimatological conditions and edaphic factors. It has not yet been clarified whether these fungi have any mycorrhizal formation; however, it is suggested that, excluding wood-decaying species, the members of Clavariaceae are biotrophic (Birkebak et al., 2013).
Handling Editor
Andrzej Szczepkowski; Warsaw University of Life Sciences – SGGW, Poland; https://orcid.org/0000-0002-9778-9567