. Introduction
Gene flow between and among populations promotes genetic diversity. The idea was expressed long ago in Wright’s (1943) isolation by distance model, where gene flow is dependent on the distance between individuals and their ability to disperse propagules or pollen. A special case of gene flow is introgression: the incorporation of alleles from one taxon mixed with another in generations after interspecific hybridization has occurred (Ellstrand, 2014).
Natural hybridization events between diploid and tetraploid species are generally phenomena rarely confirmed in nature, as contrasting ploidy levels represent a known, highly effective barrier to hybridization (Brown et al., 2023; Husband & Sabara, 2004; Koutecký et al., 2011). Abnormal endosperm ratios of maternal to paternal genomes at fertilization prevent hybrid seed formation and later hybrid sterility, leading to aneuploid gametes. However, in some genera, this phenomenon is relatively common, such as in Capsella (Han et al., 2015) and Epidendrum (Pinheiro et al., 2010). In the case of a triploid F1 generation that is formed, its gametes, both haploid and diploid, can serve as vectors for further introgression (Henry et al., 2005). Introgression from a lower to a higher ploidy level is generally referred to as the triploid bridge mechanism (De Storme & Mason, 2014; Husband, 2004; Mallet, 2007). It seems that a female triploid bridge, through unreduced egg cells, is the main pathway underpinning polyploid formation, as illustrated for Ranununculus kuepferi (Schinkel et al., 2017) and other species (Mallet, 2007; Nemorin et al., 2013). This was also documented among Aconitum species by genetic markers (Sutkowska et al., 2017a) and cytogenetic markers (Zieliński, 1982a,b). A triploid bridge could occur in both ways: from diploid to tetraploid or from tetraploid to diploid. A reversed triploid bridge was documented for Aconitum species (Sutkowska et al., 2017a), Betula (Thórsson et al., 2001), Dactylorhiza (Naczk et al., 2015), and Galeopsis (Bendiksby et al., 2011).
The plant genus Aconitum is an interesting object of population genetic studies because it shows both specific and interspecific gene flow. This genus forms a natural hybrid zone in sympatric zones of its native geographical distribution in the Carpathians (Sutkowska et al., 2013a, 2017a). The occurrence of a spontaneous diploid hybrid species has been documented (Mitka & Starmühler, 2000). Additionally, several taxonomical hybrids within the tetraploid species, as well as between diploids and tetraploids, have been described over the years (see ww.ipni.org; Mitka, 2003; Mitka et al., 2021). In studies of Aconitum species in the Sudetes and Carpathians, evidence for historical reticulate evolution and interspecific introgression was found (Mitka et al., 2007, 2015).
The role of topographic differences in the formation of variability and gene flow is commonly known (e.g., Abdelaziz et al., 2021). In our previous study on the genetic structure of marginal populations of Aconitum bucovinense in the Western Bieszczady Mountains (Eastern Carpathians), two relic populations of the species in remote peaks, separated from each other by a 10 km distance, were strongly genetically differentiated (Boroń et al., 2011). The evidence for this phenomenon is provided by numerous phylogeographic studies, especially when the occurrence of the species is connected with a distinct habitat (e.g., Stachurska-Swakoń et al., 2013, 2020).
In mountainous regions, Aconitum species are usually found along streams in deep valleys, where their populations are isolated by ridges and mountain peaks. In the previous study (Sutkowska et al., 2017a), we found introgression between Aconitum in a single mountain valley. In this study, we explored the extent of gene flow between geographically isolated Aconitum populations located in adjacent mountain valleys. We expected that gene flow between the valleys would be hindered by these topographic barriers. The second objective was to determine the genetic variability of the species in the valleys studied, with particular emphasis on hybrids. In addition, we sought to identify potential cryptic introgression and hybrids (without morphological signs of mixed ancestry) in these mountain populations of Aconitum. To address these objectives, we conducted research on the genetic variation of three species of Aconitum: A. variegatum, A. lasiocarpum, and A. firmum, as well as two hybrids: A. ×berdaui and A. ×pawlowskii from two isolated populations in the Tatra Mountains, the highest mountains in the Carpathians.
The nomenclature and taxonomy of Aconitum follow that of http://www.ipni.org, Mitka (2003), Mitka et al. (2021), Novikoff and Mitka (2011), and Starmühler and Mitka (2001).
. Material and methods
Study species
Monkshood (genus: Aconitum; family: Ranunculaceae; order: Ranunculales) belongs to an evolutionary old plant family that emerged ca. 115 million years ago (Magallon et al., 2015). Its phylogenetic history in Europe can be dated to the Late Tertiary (Boroń et al., 2020). The Aconitum subgenus Aconitum consists of two cytotypes, diploid and tetraploid, having different ecological niches. The diploids (sect. Cammarum) are mainly lowland forest species, whereas the tetraploids (sect. Aconitum) are open-landscape high mountain species (Mitka et al., 2021). In mountainous regions, both lines meet to form a hybridization zone (Sutkowska et al., 2013a, 2017a).
The species of Aconitum are perennial plants pollinated only by bumblebees (Utelli & Roy, 2000). Klepacz-Baniak (2011) has shown that bumblebees in mountainous regions travel only short distances (at most 200–300 m from their nests), which could prevent the transfer of alleles, even between nearby populations. Furthermore, the genetic diversity of natural hybrids does not show a uniform pattern, with some hybrids showing less variability and others showing much greater variability than the parental forms (Sutkowska et al., 2013b).
The ploidy of the Aconitum species has been studied before, also from the Tatra Mts, and it is constant across the particular taxa. Aconitum subgen. Aconitum in the Tatra Mountains consists of the diploids (2n [2x] = 16) A. variegatum L. and A. lasiocarpum Gáyer subsp. kotulae (Pawł.) Starm. & Mitka, and the tetraploid (2n [4x] = 32) A. firmum Rchb. (Ilnicki & Mitka, 2009, 2011). Crossing A. firmum and A. variegatum yields a partially sterile hybrid, triploid, of A. ×berdaui Zapał. (2n [3x] = 24, see Wacławska-Ćwiertnia & Mitka, 2016; Zieliński 1982a,b). A. lasiocarpum and A. variegatum form a fertile hybrid A. ×pawlowskii Mitka & Starmühl. (Mitka, 2003; Mitka & Starmühler, 2000). Both diploids and their hybrids occur in lower mountain forest zones. In contrast, the tetraploid Aconitum firmum is a high mountain species found in subalpine and alpine zones (Mitka et al., 2021), though it occasionally can descend into mountain forest zones along torrents (Mitka, 2003). Here, it inhabits man-made glades and tallgrass communities attributed to historical pastoral farming (Stachurska-Swakoń, 2009).
A rare triploid hybrid, A. ×berdaui, is morphologically related to A. firmum (e.g., the shape of the central segment of the leaves), but the half back-bent spurs of the nectaries, slightly elongated helmet (1.4–2.0× higher than wide), and slightly divided bracteoles point to its hybrid origin. In A. firmum, the spur of the nectaries is capitate, the helmet is 1.2–1.4× higher than wide, and the bracteoles are divided. In A. variegatum, the spur of the nectaries is semi-spiral coiled, the helmet is elongated, 1.6–2.5× higher than wide, and the bracteoles are spathulate or lanceolate-ovate (Mitka, 2003; Mitka et al., 2021).
In its typical habitat, A. ×pawlowskii overlaps with A. variegatum in terms of their morphological variability; however, the former’s pedicels above the bracteoles are straight glandular pilose. In A. lasiocarpum, the pedicels are entirely glandular pilose, and in A. variegatum, they are always glabrous (Mitka et al., 2021; Mitka & Starmühler, 2000).
Study area
Mała Łąka valley (Dolina Małej Łąki, ML)
The ML valley is one of the valleys in the Tatra Mountains (Western Carpathians), located in Poland (Figure 1). It is the only valley in the Tatra Mountains that has been entirely carved out of sedimentary rock, where only a few ridges are covered with crystalline rocks. A stream appears only in the lower part of the valley (called the Małołącki stream). The valley is located at 938–1,200 m above sea level; its length is 5.4 km, and its area is 5.7 km2. The valley has no side branches, only gullies.
Figure 1
Geographic locations of the studied plant populations. Poland (A). Tatra National Park (© Geoportal) (B). A map of the Miętusia and Mała Łąka valleys in the Tatra Mountains and sampling localities (for their description, see Table 1) (C); 1 – forest, 2 – glades, 3 – subalpine/alpine thickets and meadows, 4 – Aconitum ×berdaui.
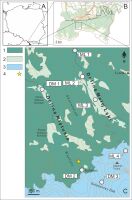
The northern part of the valley is covered with forests, and its middle part is dominated by spruce trees and meadows, forming subalpine forest vegetation. The southern part of the valley is mostly overgrown with dwarf pine trees Pinus mugo L.
Miętusia valley (Dolina Miętusia, DM)
The DM valley is located near the ML valley (Figure 1) and consists of three segments: Kobylarz, Kobylarzowy Żleb, and Niżnia Miętusia Rówień. It is a side valley and the largest branch of the Kościeliska Valley in the Polish Western Tatras. It has an area of approx. 6 km2 and a length of approx. 4.5 km; it is located at 947–1,800 m a.s.l. Almost the entire DM valley consists of sedimentary rock: dolomite and limestone. In such bedrock, typical karst phenomena develop, including underground water-flow slots, the disappearance of water, and numerous caves. The lowest part is a typical river valley. Starting from the Niżnia Miętusia Rówień (subalpine forest glade), it forms a typical glacial valley.
Kobylarz is a meadow at an elevation of about 1,430 m a.s.l., located on the eastern slopes of the DM valley. It is in a limestone area covered with rich calciphilic flora (Mirek & Piękoś-Mirkowa, 2008).
The Kobylarzowy Żleb (1,420–1,800 m a.s.l.) is a deep gorge with steep walls built of limestone, carved by a glacier in the past (Figure 1). There are numerous typical mountain plant species here, some of which grow only in the Tatra Mountains (Mirek & Piękoś-Mirkowa, 2008). Both valleys are separated by a mountain ridge called Skoruśniak (Figure 1).
Collecting of samples
Samples of Aconitum individuals (fresh leaves stored in silica gel) were collected from all local subpopulations of studied taxa that we found at 850 to 1,800 m a.s.l. in the valleys Mała Łąka (ML) and Miętusia (DM) (Figure 1). Altogether we found four subpopulations in ML and seven subpopulations in DM. The number of sampled individuals was dictated by the subpopulation size; if subpopulations were small, the leaves were collected from all possible individuals. As a result, 24 ML and 23 DM samples were collected from three species and two hybrids (Table 1, Figure 1). The taxonomical variability of the specimens representing the sampled subpopulations was identified following Mitka (2003). The vouchers of the specimens from the subpopulations are deposited in KRA.
Table 1
Aconitum specimens sampled in the Mała Łąka valley (ML) and Miętusia valley (DM) in the Tatra Mountains (see also Figure 1) (ploidy level see Ilnicki & Mitka, 2009, 2011).
DNA isolation and ISSR fingerprints analyses
DNA was isolated from fully expanded, mature, and healthy leaves. Their DNA was extracted from each sample (20 mg) using the Genomic Mini AX Plant kit (A&A Biotechnology). The concentration and purity of DNA in the samples were measured using a Nanodrop 2000C spectrophotometer (ThermoFisher Scientific). Five ISSR primers were used, whose sequences (Table 2) were obtained from Stepansky et al. (1999) and Sutkowska et al. (2013b). The PCR amplification reactions were performed on a 2720 thermal cycler (Applied Biosystems). Each amplification was carried out in a 25 µl reaction mixture: 2.5 µl of 10-fold concentrated reaction buffer supplied by the Taq DNA polymerase manufacturer (ThermoFisher Scientific), 1.5 mM MgCl2, 0.19 mM of each dNTP (ThermoFisher Scientific), 27 pmol primer, 100 ng template DNA, and 1.4 Taq polymerase units. The annealing temperature for the primers ISSR2, ISSR4, and ISSR7 was 44 °C; for ISSR1 and ISSR3, it was set to 47 °C. The optimal conditions for the reaction were as follows: initial denaturation at 94 °C for 5 min, 42 amplification cycles: denaturation at 94 °C for 59 s, annealing at 44 °C (47 °C) for 59 s, then polymerization at 72 °C for 59 s, with a final polymerization at 72 °C for 7 min. A negative control reaction without any DNA template was included in each amplification.
Table 2
Primers used in the PCR, number of reaction products generated by each primer, range number of PCR products per sample, and mean number of PCR products per Aconitum sample in the Mała Łąka valley (ML) and Miętusia valley (DM) in the Tatra Mountains.
| Primer | Primer sequence | Number of PCR products | Range | Mean specimen |
|---|---|---|---|---|
| ISSR1 | (TC)8C | 23 | 4–10 | 8 |
| ISSR2 | (AG)8T | 27 | 6–11 | 9 |
| ISSR3 | (GGGTG)3 | 19 | 4–10 | 7 |
| ISSR4 | (ATG)6 | 22 | 3–11 | 7 |
| ISSR7 | (AC)8T | 22 | 5–9 | 7 |
The obtained products were separated in 1.5% agarose gel with ethidium bromide (0.5 µg/ml) in TBE buffer at 100 V/25 cm for 90 min. The electrophoresis results were captured using Imagemaster VDS (Amersham Pharmacia Biotech Ltd.) with Liscap Capture Application ver. 1.0. Electrophoretic gel analysis was performed using GelScan ver. 1.45 (Kucharczyk Electrophoretic Techniques). The creation of a calibration curve based on the length band pattern of markers (GeneRuler TM 100 bp, ThermoFisher Scientific) enabled the determination of the molecular weight of the resulting amplification products. ISSR reproducibility tests (Bonin et al., 2004) included replicates within the plate (n = 12) and between plates (n = 9). The data have been deposited in the public repository: doi: https://doi.org/10.5061/dryad.mpg4f4r2w and https://doi.org/10.5061/dryad.pk0p2ngw7.
DNA sequencing
Total DNA extracts served as templates in the amplification of the chloroplast DNA region trnL(UAG)–ndhF with primers trnL(UAG) – 5′-CTGCTTCCTAAGAGCAGCGT-3′ and ndhF – 5′-GAAAGGTATKATCCAYGMATATT-3′ (Shaw et al., 2007). The reaction was carried out in a total volume of 50 µl that contained 1× DreamTaq Green buffer (ThermoFisher Scientific); 3.5 mM MgCl2; 0.08 mM of dNTPs; 0.08 µM of each primer; and 1 µL of DreamTaq DNA polymerase (ThermoFisher Scientific). The amplifications were conducted on a T100 Thermal Cycler (Bio-Rad), using this temperature profile: 5 min of initial denaturation at 94 °C; 25 touchdown cycles composed of 30 s each at 94 °C; 30 s of decreasing annealing temperature (0.5 °C/cycle from 67.5 °C in the 1st to 55 °C in the 25th cycle); 1 min at 72 °C; and 20 cycles each of 30 s at 94 °C, 30 s at 55 °C, and 1 min at 72 °C; and ending with 10 min at 72 °C for the final extension step. Amplification efficiency was verified by agarose gel electrophoresis, and the positive PCR products were purified with the Clean-Up DNA purification kit (A&A Biotechnology). For the purified PCR products, the fragment of the trnL(UAG)–ndhF amplicon was sequenced using the internal primer V2_F – 5′-GTTCGCAAAGAACTGAAGTGAC-3′ (Sutkowska et al., 2017a). Sequencing was done with a BigDye Terminator v3.1 Cycle Sequencing Kit (Life Technologies) on a T100 thermal cycler (Bio-Rad) and a 3500 Series Genetic Analyzer (Life Technologies) by applying standard protocols. The obtained sequences were processed and aligned by MEGA 6 software (Tamura et al., 2013), followed by their manual adjustments. All generated sequences have been deposited in GenBank (https://www.ncbi.nlm.nih.gov/nuccore/) with these accession numbers from KU314873-KU314895.
Data analysis
At the population level, we analyzed PCR-ISSR polymorphism as alleles under the following assumptions: ISSR products segregate as dominant alleles in Mendelian fashion, genotype frequencies at ISSR loci are in Hardy-Weinberg equilibrium, and comigrating fragments are considered homologous loci (Apostol et al., 1996). STRUCTURE (Pritchard et al., 2000) was used to identify genetically homogeneous groups of individuals; this software tool places individuals in K clusters characterized by a distinct set of allele frequencies at each locus. An admixture ancestry model was fitted, and allele frequencies were correlated; 3 × 105 of Markov chain Monte Carlo (MCMC) replicates, with a burn-in of 5 × 104 iterations, were checked from K = 2 to K = 9; the K = 4 parameter was in line with the results of the taxonomic analysis.
Table 3
AMOVA results for the Aconitum species in two valleys in the Tatra Mountains based on 161 ISSR loci.
| Source of variation | Sum of squares | Variance components | Percentage variation* |
|---|---|---|---|
| Among species | 190.978 | 3.970 | 17.404 |
| Within species# | 791.283 | 18.840 | 82.595 |
| Species between valleys | 147.990 | 6.102 | 24.804 |
| Among species within valleys | 154.701 | 4.851 | 19.717 |
| Within species | 395.823 | 13.649 | 55.479 |
The matrix of samples and ISSR products, coded as binary (0/1) data, was used to calculate the Nei and Li (1979) distances and the Neighbour Net (NN) ordination in SplitsTree ver. 4.12 software. Bootstrap values (BS) for the clusters were determined based on 1,000 random runs. Additionally, the Nei and Li distances were used for the principal coordinate analysis (PCoA) and to derive the minimum spanning tree (MSP). Calculations were made using the NTSYSpc ver. 2.11 multivariate analysis package (Rohlf, 2002).
Hierarchical analysis of molecular variance (AMOVA) was implemented using the Arlequin ver. 3.11 program (Excoffier et al., 2005).
. Results
The PCR-ISSR analysis resulted in 161 polymorphic loci. Data quality testing indicated high repeatability across the ISSR bands, being greater than 97%. The total number of PCR-ISSR bands generated by the primers varied from 19 to 27, and the number of bands generated by the individual primers per sample varied from three to eleven, with a mean of seven to nine (Table 2). A. lasiocarpum was the only one that had private alleles.
Taxonomically, in the ML valley occurred A. firmum (one in collected specimens), A. ×pawlowskii (9), A. variegatum (3), and A. lasiocarpum (11). In the DM valley, A. firmum (8), A. ×pawlowskii (7), and A. variegatum (7) were identified, and only one individual of A. ×berdaui was found (Figure 1, Table 1).
Sequencing of the trnL(UAG)–ndhF region of cpDNA revealed the presence of two types of Aconitum cpDNA haplotypes, as described in our previous articles (Boroń et al., 2020; Mitka et al., 2016; Sutkowska et al., 2017a). They are distinguished by two unique indels and form a type I in tetraploids and type II in diploids (Boroń et al., 2020). The hybrid A. ×berdaui has a type I cpDNA haplotype.
The percentage variation between species, irrespective of the valley, was lower (17.4%) than the genetic partition of species between the two valleys (24.8%, Table 3). This means that the species in the neighboring mountain localities DM and LM had different ISSR genetic profiles, which was further confirmed by the PCoA and NN analyses.
The Aconitum taxa in the two valleys were genetically differentiated (Figure 2, Figure 3). The first three axes of PCoA explained 32.79% of the total genetic variability of ISSR. In the ordination diagram, the two valleys DM and ML separated along PCoA 1 (Figure 2); PCoA 2 discriminated A. lasiocarpum and PCoA 3 A. firmum in the ML valley, while A. firmum in the DM valley layed close to the diploid group. The hybrid A. ×berdaui was placed in a separate position among the tetraploid A. firmum along PCoA 3 and evidently genetically close (minimum spanning tree) to the A. ×pawlowskii APK7 sample (Figure 3).
Figure 2
Principal coordinate analysis (PCoA, Dice distance, NTSYSpc) of 47 specimens of Aconitum in two valleys in the Tatra Mountains: Mała Łąka (A) and Miętusia (B), based on 161 ISSR markers. The percentages in the axis titles are the proportion of explained variation.
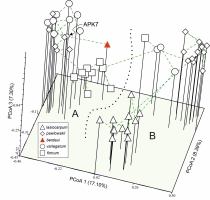
Figure 3
Classification (Neighbour Net, NN, SplitsTree) of Aconitum species in Miętusia (DM, left) and Mała Łąka (ML, right side of the diagram) valleys in the Tatra Mountains and STRUCTURE grouping (K = 4, pie diagrams), based on 161 ISSR bands. Bootstrapped Nei and Li distances based on 1,000 random runs. A. ×berdaui ABK1 is introgressed with individual A. ×pawlowskii APK7. For the sample abbreviations, refer to Table 1.
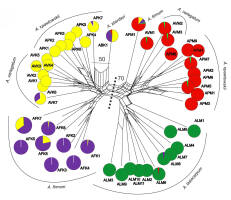
A main split (BS 70%) in the NN ordination distinguished the genetic stocks of the Aconitum species in the two valleys. In DM 2, the triploid A. ×berdaui ABK1 was genetically close to diploid A. ×pawlowskii APK7 (BS 50%). The NN ordination confirms the origination of the hybrid A. ×berdaui from tetrapoid A. firmum and diploid complex A. variegatum/pawlowskii (Figure 3).
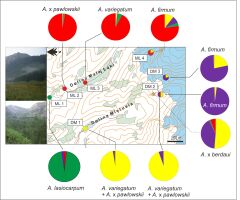
According to the STRUCTURE results, evidence for the possible interspecific introgression was found between the diploid complex and the tetraploid A. firmum. In DM 3, a mixed ancestry was found in A. firmum AFK7 and AFK8. Similarly, A. firmum AFM1 in the ML 4 locality was of mixed ancestry and featured the genetic stocks of the diploid complex from both valleys, as well as A. firmum from DM 3. All the introgessed A. firmum cases were located above the tree line (Figure 4).
Figure 4
Results of STRUCTURE (K = 4) of Aconitum species in two valleys in the Tatra Mountains. Pie diagrams display the share of particular DNA-ISSR Bayesian groups in each sampled locality. Open areas among forest (green) and above tree line (blue) are demarcated. Photo insert: upper, Mała Łąka at lower elevations; bottom, Miętusia, in the foreground Kobylarzowy Żleb segment at treeline (photo: T. Zagórski). For detailed results of the STRUCTURE analysis, refer to Figure 3.
Furthermore, a portion of the A. firmum genome can be found in the diploids A. variegatum AVK7 and A. ×pawlowskii APK7 (Figure 3). Introgressed A. variegatum AVK7 was located at a lower elevation in the DM 1 population, and A. ×pawlowskii APK7 co-occurred with A. firmum and A. ×berdaui near the tree line in DM 2. A sample of A. lasiocarpum ALM8 in the low-lying population ML 1 was introgressed with A. firmum (Figure 3, Figure 4).
The numerical analyses of ISSR markers confirm that gene flow occurred between studied specimens of the diploid A. lasiocarpum and the diploid A. variegatum and A. ×pawlowskii. It was found in all samples of A. variegatum (AVM1-3) in ML 3 and in two samples of A. ×pawlowskii (APM5 and APM7) in ML 2 located at lower elevation in the valley. One sample of A. lasiocarpum (ALM8) had part of the diploid complex (A. variegatum and A. ×pawlowskii) in ML 1 (Figure 3, Figure 4).
. Discussion
Genetic diversity between populations
In this study, the STRUCTURE clustering revealed three genetic groups within the diploids: A. lasiocarpum and the complex A. ×pawlowskii/variegatum, being differentiated across the valleys. Moreover, a significant difference in genetic variance partitioning (AMOVA) was found between the species populations in the two valleys studied and corroborated by the NN clustering dendrogram, where the distinction was supported by a relatively high BS (70%). The division between valleys is expressed by the PCoA (axis 1), and this analysis with the distribution of Aconitum samples along axis 1 indicated different genetic profiles of the species. It could be a result of the separate genetic profile of A. lasiocarpum that occurred in only one of the valleys. Restricted or no gene flow between the valleys can be attributed to a topographical barrier, with the mountain peaks and ridges that separate the valleys making the movement of diaspores and pollen difficult, or even impossible, between them (Mitka et al., 2022). However, these barriers gradually disappear at higher elevations where both studied valleys flatten, reducing the difference in height between their bottoms and separating peaks, such that gene flow becomes possible. This is evidenced by the structure of the genomes of samples located at higher elevations (above 1,350 m), including that of A. firmum (AFM1) and its hybrid A. ×berdaui (ABK1) which has genomes from both valleys.
One of the particular results is the close genetic relations of A. variegatum and its hybrid A. ×pawlowskii and their clear differentiation between adjacent valleys. While their close genetic affinity is understandable from a taxonomic point of view, their differentiation between valleys is unexpected. In our earlier study of two endemic tetraploid species, A. firmum and A. bucovinense, and their hybrid A. ×nanum in the Carpathians a greater percentage of their variation is attributable to the ISSR genetic groups (22.6%) than to the taxa (15.6%, all p < 0.001, AMOVA; Sutkowska et al., 2017b). It means that genetic ISSR-fingerprinting is not relevant in strict taxonomic studies, but it is a sensitive tool for population studies on a local scale.
Genetic diversity in populations
Generally, in mountain valleys, various intrinsic dynamic processes, such as fluctuations in population sizes due to bottlenecks, invasions, founding events, and ecological succession (Avise & Hamrick, 1996; Stachurska-Swakoń et al., 2011, 2020), can all affect the genetic diversity of plant populations. The complexity of mountains is tightly associated with high biodiversity (Perrigo et al., 2020).
Among the examined populations, the ML valley has the highest genetic disparity between species, being affected by the presence of a genetically distinct A. lasiocarpum in this valley. Because the latter presents a genetically consistent population, it suggests a bottleneck/founder effect. Moreover, A. lasiocarpum was the only species that had private loci and, thus, was the most genetically discriminated. In the case of the encountered hybrid A. ×pawlowskii, it had no additive (parental) ISSR loci. An additive contribution of the genomes of two parental species has been uncovered, for example, in natural hybrids between the forest grass species Bromus benekenii and B. ramosus (Sutkowska et al., 2015) and likewise, Phoenix canariensis and P. dactylifera (Gonzalez-Perez et al., 2004). Similar results were obtained while studying two fir species in the narrow contact zone at an elevation of 1,000–1,100 m in Japan, where nine of 78 tree individuals were genetically intermediate and natural hybrids, whose genetic analysis confirmed they had backcrossed with parental species; it turns out that the hybrid species of intermediate morphology had previously taxonomic independent status (Aizawa & Iwaizumi, 2020). The lack of additivity in A. ×pawlowskii was reported for Aconitum populations in another mountain valley in the Tatra Mountains (Sutkowska et al., 2017a), as well as in a natural hybrid zone in the Beskid Niski (Western Carpathians, Sutkowska et al., 2013a). In both studies, the hybrid species have a specific genetic makeup that differs from that of either parent species. In the current study, the lack of additive genetic effects in A. ×pawłowskii may be the result of asymmetric introgression between this species and its parent species. This explanation was postulated before in a study of the hybrid zone of A. lasiocarpum and A. variegatum in the Western Carpathians (Sutkowska et al., 2013a). This mechanism could also be responsible for the genetic similarity between the hybrid and one of its putative parents. However, fully resolving the problem will require further studies entailing an array of genetic markers.
Hybridization and introgression
In the valleys, in addition to pure Aconitum species, the hybrids A. ×pawlowskii and A. ×berdaui (the latter only in the DM valley) were found. Their hybridogenous status was proven by morphological key characters (see Mitka et al., 2021). Their mixed ancestry was also revealed by ISSR markers in the previous studies (Sutkowska et al., 2013a). All diploid and tetraploid genetic introgressants were taxonomically (morphologically) pure specimens, a phenomenon also found in other studies (Beatty et al., 2015; McIntosh et al., 2014).
We know that A. ×pawlowskii originates only in the contact zone of the parental species (Sutkowska et al., 2013a). In the present study, A. ×pawlowskii was not conclusively shown to be a genetic hybrid, despite its taxonomic designation, because the ISSR profile was very similar to its putative parent, A. variegatum. The small outcome of genetic additivity could be an effect of introgression on a local geographical scale. This phenomenon seems common in hybridizing plant species (Tagane et al., 2018). It could be attributable to differential introgression in a hybrid zone where many loci have gradual changes in allele frequency and do not represent regions of the genome that define species boundary (Harrison & Larson, 2014).
Bayesian inference indicates that maximum levels of gene flow occur at the intrapopulation level for all plant populations sampled in that study. The detection of diploid- and tetraploid-specific markers in the trnL(UAG)–ndhF region of cpDNA allows for the inference of the introgression direction between diploid and tetraploids through a triploid bridge (Sutkowska et al., 2017a).
According to the STRUCTURE analysis, in both valleys studied, A. firmum was strongly introgressed with ISSR markers typical for diploid species. The two samples of A. firmum (AFK7 and AFK8) contain fragments of the nuclear genome belonging to the diploid A. ×pawlowskii/variegatum. This result suggests a mixed ancestry of AFK7 and AFK8, one composed of ISSR genetic groups typical for different ploidy levels. That A. firmum AFM1 in ML 4 has a complicated genetic ancestry; it is due to its genome being partly composed of both diploid and alien tetraploid genetic ISSR groups. It is likely that part of the diploid genome could be transferred from the neighboring open-site population, introgressed with the tetraploid genome of A. firmum in DM 3.
Such a mixed diploid/tetraploid ancestry could be an effect of introgression via the triploid bridge mechanism. These cryptic introgressants did not differ morphologically from the pure species. As previously reported, the degree of introgression in populations is not always reflected in the morphology of species (Čertner et al., 2020; Sutkowska et al., 2013a). However, when introgression of cpDNA occurs from a tetraploid to diploid species – a case not found in the present paper – the bracteoles of the diploid have a shape that is typical of the tetraploid (Sutkowska et al., 2017a).
All the analyses, Bayesian inference (STRUCTURE), NN, minimum spanning tree, and PCoA, concurred in showing that A. ×berdaui ABK1 had two different genomes: one was derived from tetraploid plants and the second from diploid plants (Figure 5). In diploid A. ×pawlowskii APK7, we found a tetraploid part of the nuclear genome. We infer that APK7 was introgressed via a reversed triploid bridge (i.e., from the tetraploid to the diploid line). Similarly, the diploid A. variegatum AVK7 was introgressed with A. firmum, as well as A. lasiocarpum ALM8. On the other hand, the tetraploids A. firmum AFK7 and AFK8 introgressed with the A. ×pawlowskii/variegatum complex, similar to AFM1, which was introgressed with both A. ×pawlowskii/variegatum and A. lasiocarpum. A similar result was found in a previous study (Sutkowska et al., 2017a), where the flow of nuclear DNA was coupled with the transfer of maternal cpDNA from the tetraploid to diploid species (i.e., a reversed triploid bridge). In our study here, given that we did not find a transfer of the plastid marker between the diploid and tetraploid species, we designate the mechanism as a cryptic reverse triploid bridge. Thus, the triploid was probably only a pollen donor. Forty years ago, Zieliński (1982a,b) reported that the triploid A. ×berdaui can produce 13% of viable pollen in the Tatra Mountains, and such triploid pollen could cross both diploid and tetraploid lines (Ramsey & Schemske, 1998). Hence, we may assume that the transfer of genome fragments from the tetraploid to the diploid was carried out by the paternal line.
Figure 5
Scheme representing the different observed pathways of backcrossing in the diploid and tetraploid lines of Aconitum. (a) The triploid formation is depicted as the first step of a triploid bridge; (b) Cryptic triploid bridge (diploid – triploid – tetraploid); (c) reversed triploid bridge (tetraploid–triploid–diploid); – not observed.
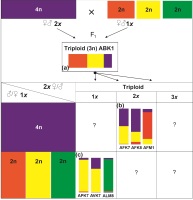
We propose the following scenario. The initial donor of maternally inherited plastid DNA in the hybrid triploid was the tetraploid A. firmum. Subsequent backcrossing involved monoploid maternal gametes (n = 1x) from a triploid line (3x) and normal paternal pollen from a diploid line (2x): A. lasiocarpum, A. variegatum, or A. ×pawlowskii (scenario a, Figure 5). In the second step, maternally inherited cpDNA was introduced from the tetraploid to diploid species after backcrossing the triploid (1x ♀ gamete) with the parental diploid (1x ♂ gamete) (scenario c, Figure 5). In this way, tetraploid-specific cpDNA could be introgressed to diploid species via a reversed triploid bridge mechanism (Sutkowska et al., 2017a).
Based on the findings of the present study, we hypothesize that the union of gametes between diploid and tetraploid species first led to the formation of a triploid. In this respect, the introgressed individuals are represented by A. variegatum: AVK7, A. ×pawlowskii APK7, and A. lasiocarpum: ALM8 (Figure 3, Figure 4).
This work also revealed the introgression of diploid species (A. variegatum, A. lasiocarpum, or their hybrid A. ×pawlowskii) to the tetraploid A. firmum. Accordingly, the result of that introgression is present in AFK7, AFK8, and AFM1 (Figure 3).
. Conclusions
The crossing between species that possess different levels of ploidy with distinct ecological profiles is rarely observed in nature. In the studied plant populations, we found evidence for introgression among diploid and tetraploid Aconitum species. The triploid species A. ×berdaui played a crucial role in this process. This study confirms our previous findings that a hybrid triploid can be a donor of male and female gametes, although the latter seems to be an exceptional phenomenon. Morphologically, species in the valleys of the Tatra Mountains are often of mixed ancestry and form cryptic hybridogenous forms. More importantly, populations of the same species in adjacent valleys differ genetically. These pronounced differences could be the outcome of a complex effect of various mechanisms, including founder effects, bottlenecks, and restricted gene flow between populations. Notably, the flow of genes within the same ploidy level is restricted to one valley and to neighboring populations, and most of the introgressive forms grow near or above the tree line. The subpopulations of the Aconitum genus in the Tatra Mountains form genetically differentiated units. The restricted gene flow could be among the mechanisms able to promote their genetic variability.
. Ethics
We did not cause any form of harm to the studied species. Our research does not include studies on human subjects, human data, human tissue, or animals. Permission: no. DNE.404/145/13.
. Supplementary material
All data, code, and electronic supplementary materials are publicly available on Dryad: https://datadryad.org/stash/share/OEXi-2bIwFYUbi9r1FVuD3pbw4mfyJNfc_EyWShqIlI
doi: https://doi.org/10.5061/dryad.mpg4f4r2w and https://doi.org/10.5061/dryad.pk0p2ngw7


