. Introduction
The Kurdistan Region in the Iraq highlands in the northeast of the country is the only place in Iraq where there are natural forests (Nasser, 1984). The highlands are made up of a series of mountains that stretches from Turkey’s north to Iran’s adjacent east. Approximately 90% of the total forest cover is made up of oak forest, with the remaining 10% consisting of other wood (Guest & Al-Rawi, 1966; Khwarahm, 2020).
Quercus aegilops is the most common species of oak, followed by Q. libani, Q. infectoria, and Q. macranthera. The remaining species, with the exception of Q. macranthera, are regarded as native to the area (Nasser, 1984; Zohary, 1973). About 70% of the oak woodlands are made up solely of Q. aegilops (Shahbaz, 2010).
The term “endophytic fungi” refers to fungi that reside in plant tissues for all or a portion of their life cycle, developing a mutually beneficial symbiotic relationship with their host plant while avoiding causing negative effects or pathogenicity (Rodriguez et al., 2009). Various plant parts, including the bark, flowers, fruits, leaves, roots, stem, and scales, have been found to have endophytes (Pirttilä et al., 2008). According to recent studies, there are probably more than 3 million species on the planet, of which only about 150,000 have been described. However, it is currently thought that there are about a million different kinds of fungi that are endophytes (Bhunjun et al., 2024; Hyde et al., 2020; Phukhamsakda et al., 2022).
Endophytes help plants in a number of ways, including promotion of growth, defense against diseases or pests, assistance with phosphorus uptake, and increasing plant tolerance to biotic and abiotic stressors (Hardoim et al., 2015; Schulz & Boyle, 2005). Plant-associated fungi, specifically endophytic fungi, represent a highly diverse category of microorganisms, and nearly all plant species on the earth harbor endophytic fungus. They colonize interior tissues of plants without exhibiting disease signs. As a result, the host plant gains from their relationships e.g. protection against biotic and abiotic stresses, induced host resistance, and production of secondary metabolites (Baron & Rigobelo, 2022), while endophytes also gain from host plants, as they receive their essential nutrients, are shielded from harsh external environments, and face less competition from other microorganisms (Kumari et al., 2023).
Alternaria is a genus of fungi that includes both pathogenic and saprophytic weak facultative parasites and endophytic species. The genus Alternaria Nees is a diverse genus classified in the family Pleosporaceae, order Pleosporales, class Dothideomycetes, and phylum Ascomycota of the kingdom Fungi. Recent taxonomic revisions of the genus based on multilocus phylogeny divided the genus into 29 taxonomic sections (Bessadat et al., 2021; Lawrence et al., 2016; Li et al., 2023), and nearly 370 species are currently recognized as belonging to the genus (Bessadat et al., 2021; Li et al., 2023; Wijayawardene et al., 2020).
Endophytic Alternaria species are capable of producing a diverse range of secondary metabolites. Inhibitory effects on pathogenic bacteria have been demonstrated for a number of bioactive chemicals that have been discovered in endophytic Alternaria species and have shown promise in the treatment of human diseases like cancer, diabetes, and HIV (Eram et al., 2018).
Potential phytotoxins produced by Alternaria have biotechnological applications as mycoherbicides or biocontrol agents for a wide range of plant species in different habitable zones (Costa et al., 2020; Lawrence et al., 2016; Woudenberg et al., 2013, 2015). Additionally, Alternaria species also produce mycotoxins that contaminate feed and food commodities and are connected to opportunistic animal and human diseases that have a serious impact on patient health (Meena et al., 2017; Tralamazza et al., 2018). Alternaria alternata and A. infectoria have been regularly reported as causative agents of phaeohyphomycosis in patients undergoing kidney transplants and those with immune impairment (Cardona et al., 2020; Lopes et al., 2013).
The goal of the present study was to isolate and characterize endophytic Alternaria species associated with two Quercus species (Q. aegilops and Q. infectoria) from Iraq based on morphological and DNA sequence data and to reconstruct their phylogenetic relationship.
. Material and methods
Study site
The study was conducted during September–November 2020 in the Duhok province, which is located in the northwest of the Kurdistan Region, Iraq. It is located 433 to 1,512 m above sea level, between latitudes 36°18′ and 37°20′ N and longitudes 42°20′ and 44°17′ E. The rainy season lasts from November to March, whereas the summer months of June through September have little to no rainfall. The weather in Duhok is influenced by the Mediterranean climate; there are typically dry and hot summers and cold, wet winters. It rains on average between 500 and 1,000 millimeters every year. The summer temperature range is 20 °C to 37 °C, and the wintertime temperature range is 0 °C to 15 °C (Mzuri et al., 2022).
Sample collection and fungal isolation
Thirty samples from each of leaves and buds were collected from each of the two oak plants (Quercus aegilops and Q. infectoria), stored in sterile paper envelopes, transported to the mycology laboratory of the Department of Biology at the University of Zakho, and processed within 48 h. In the laboratory conditions, the leaves were cut into small pieces with a 5 mm diameter paper puncher; the buds were cut into small slices. The samples prepared from each plant were taken separately and sterilized at an ethanol concentration of 70% for 60 s, 3% sodium hypochlorite for 3 min, and then sterilized again with an ethanol concentration of 70% for 60 s and finally washed with sterile distilled water three times for 1 min. In a laminar air flow bench, the surface-sterilized samples were allowed to dry on sterile paper towels. The samples were transferred to Petri dishes (5 segments per plate) containing Malt Extract Agar (MEA) medium (Himedia laboratories, India) supplemented with 50 µg/mL ampicillin and streptomycin to inhibit bacterial growth and incubated at 25 ± 2 °C until fungal growth became visible (Martins et al., 2016). For the establishment of pure cultures, hyphal tips growing out from the tissue pieces were taken with a sterilized needle, subcultured onto fresh MEA plates, and incubated at 25 ± 2 °C. The growth of fungal colonies and sporulation in the cultures were observed after two weeks of incubation.
Morphological identification of Alternaria species
Identification of Alternaria isolates from pure cultures grown on PDA medium was based on some cultural and morphological characters, such as conidial dimensions, shape, color, ornamentations, and number of transverse and longitudinal septa. Fungal isolates were identified with the aid of several relevant taxonomic references (Ellis, 1971, 1976; Li et al., 2023; Simmons, 1986, 2007; Wanasinghe et al., 2018; Zhang & Zhang, 2006).
Molecular identification of Alternaria species
DNA extraction
Pure colonies of Alternaria isolates were selected and transported to a ceramic mortar to be ground into powder using liquid nitrogen. The powder was then transferred to sterile tubes and preserved in the freezer until used for DNA extraction. DNA of the isolated fungi was extracted using the Add Prep Genomic DNA Extraction Kit (Korea®;), according to the manufacturer’s instructions.
A Thermo Scientific Nano drop 2000c spectrophotometer was used to quantify and evaluate the purity of the extracted DNA in accordance with the user manual’s recommendations. The extracted fungal DNA was amplified using the universal primers ITS1 (5′-TCC GTA GGT GAA CCT GCG G-3′) and ITS4 (5′-TCC TCC GCT TAT TGA TAT GC-3′) as well as LROR (5″-ACCCGCTGAACTTAAGC-3″) and LR5 (5″-ATCCTGAGGGAAACTTC-3″), according to White et al. (1990) and Vilgalys and Hester (1990), respectively.
DNA amplification using Polymerase chain reaction
Single plex PCR and Semi-nested PCR were carried out for targeting both ITS and LSU genes, respectively. They were performed for amplification in a total volume of 45 µl reaction tube containing a mixture of 15 µl Crystal Hot Start DNA Master Mix (0.2 mM of dNTP, 1× Ex Taq Buffer and 2.0 mM of MgCl2), 1.5 µl forward primer (10 pmol), 1.5 µl reverse primer (10 pmol), 3 µl of DNA samples as a template, and 21 µl of nuclease free water for each of the primers. A thermocycler was used for amplification using the following settings for the ITS1 and ITS4 genes Using the Single plex PCR technique: 95 °C for 3 min followed by 35 cycles of 94 °C for 40 s, 52 °C for 1 min, and 72 °C for 1 min, and then final strand elongation at 72 °C was done for an additional 10 min (White et al., 1990).
In turn, the Semi-nested PCR technique amplification for LSU was as follows: 95 °C for 3 min followed by 30 cycles of 94 °C for 40 s, 67 °C for 1 min, and 72 °C for 1 min, and then final strand elongation at 72 °C was done for an additional 10 min (Al-Bedak et al., 2018). The PCR products were subjected to Agarose gel electrophoresis (1.5%) after staining with Red Safe Dye with green fluorescence (GeNet Bio, Korea). Electrophoresis was run at 80 V for 45 min. The DNA ladder with molecular weight (100–1,000 bps) was added for estimating the band size (Verkley et al., 2013). The sequences retrieved were uploaded to GenBank.
Phylogenetic analysis
Phylogenetic analysis was conducted based on both the ITS and LUS gene data using neighbor joining (NJ) approaches. The phylogenetic tree was aligned using the Muscle method associated with substitution models (K2: Kimura 2 parameter). Alignment gaps were treated as missing data. NJ trees were constructed based on the total character differences, and bootstrap values were calculated from 1,000 replications. MEGA X software 10.1.6 was used to conduct evolutionary analysis (Kumar et al., 2018).
. Results and discussion
Morphological characterization of Alternaria species
Based on morphological characters, seven endophytic Alternaria species were identified. These include 4 species in section Alternaria, 1 species in section Chalastospora, and 2 species in section Ulocladioides (Table 1). Pure cultures from representative strains of the reported species have been deposited at the culture collection of the mycology laboratory at the University of Zakho.
Table 1
Conidial morphology of endophytic Alternaria species isolated from Quercus species and grown on PDA medium.
Alternaria spp. assigned to A. sect. Alternaria
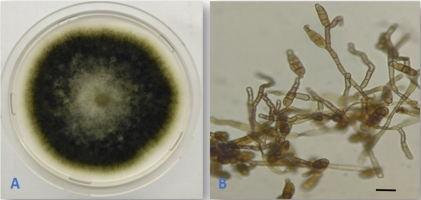
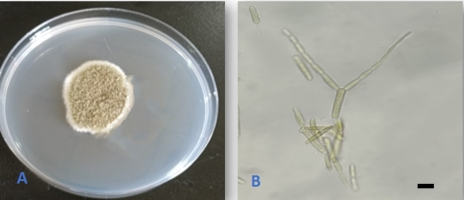
Alternaria alternata (Fr.) Keissl., Beih. Bot. Centralbl., Abt. 2, 29: 434. 1912. (Figure 1).
This is a very common species with worldwide distribution. The species was previously reported as an endophyte from several host plants (Rashmi et al., 2019). The fungus was reported as an endophyte from buds and leaves of Q. cerris in Italy (Ragazzi et al., 2001), from Q. macranthera and Q. brantii in Iran (Ghasemi et al., 2019; Ghobad-Nejhad et al., 2017), and from twigs of Q. rubra in the Czech Republic (Novotný, 2022). However, this is the first isolation of the fungus as an endophyte from Q. aegilops.
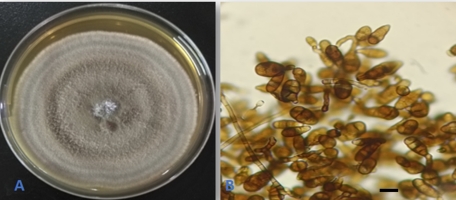
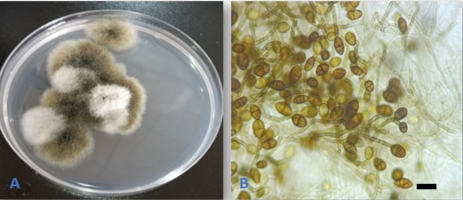
Alternaria angustiovoidea E.G. Simmons, Mycotaxon 25(1): 198(1986). (Figure 2).
The fungus was first described by Simmons (1986) based on isolates from leafy spurge (Euphorbia esula) in Canada and the USA. A. angustiovoidea was also isolated from Egyptian soils (Abdel-Sater et al., 2020). The fungus was isolated as an endophyte from leaves and stems of Suada microphylla (Sun et al., 2011) and from two Pinellia species (Kong et al., 2023). This is the first report of the fungus as an endophyte from Q. aegilops. Moreover, the species represents a new addition to the Iraqi mycobiota.
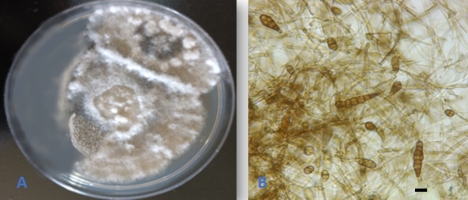
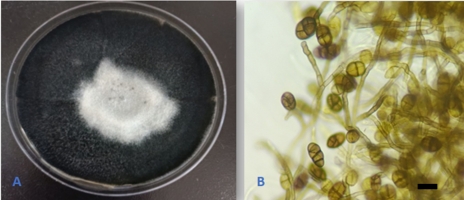
Alternaria doliconidium J.F. Li, Camporesi & K.D. Hyde, in Wanasinghe et al., Fungal Diversity: 10.1007/s13225-018-0395-7, [147] (2018) (Figure 3).
The fungus was isolated and described from spines of Rosa canina in Italy (Wanasinghe et al., 2018). This is the first report of the fungus as an endophyte from Q. aegilops and in the world. In addition, the species is reported for the first time in Iraq.
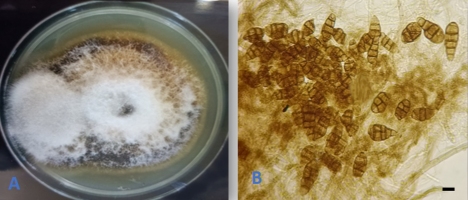
Alternaria tenuissima (Nees & T. Nees : Fr.) Wiltshire, Trans. Brit. Mycol. Soc. 18: 157. 1933. (Figure 4).
The endophytic nature of the fungus was known on a number of host plants (Rashmi et al., 2019). However, our isolation represented the first report of the fungus as an endophyte on Q. infectoria from Iraq.
An endophytic isolate of A. tenuissima was reported as a natural source of bioactive compounds that showed inhibitory activity against both gram negative and gram positive bacteria as well as against human pathogenic Candida albicans yeast and several plant fungal pathogens (Chatterjee et al., 2022).
Alternaria spp. assigned to A. sect. Chalatospora
Alternaria malorum (Ruehle) U. Braun, Crous & Dugan, Mycol. Progr. 2: 5. 2003. (Figure 5).
According to Shipunov et al. (2008), the fungus was reported as an endophyte on knapweed (Centaurea stoebe) and from Q. brantii in Iran (Alidadi et al., 2018). This is the first report of the fungus as an endophyte on Q. aegilops and represents a new addition to the Iraqi mycobiota.
Alternaria spp. assigned to Section Ulocladioides
Alternaria consortialis (Thüm.) J.W. Groves & S. Hughes [as “consortiale”], Canad. J. Bot. 31: 636. 1953. (Figure 6).
The fungus was previously reported under the name Ulocladium consortiale as an endophyte of Ziziphus spina-christi and Z. hajanensis from Oman (El-Nagerabi et al., 2013) and in Q. brantii from Iran (Alidadi et al., 2018). This is the first report of the fungus as an endophyte on Q. aegilops. The species was recently reported from Iraq on wheat grains and bran (Fadhil et al., 2022).
Alternaria sorghi (X.G. Zhang & T.Y. Zhang) Gannibal & D.P. Lawr., Mycotaxon 133(2); 296(2018). (Figure 7).
The species was originally described from leaves of Sorghum bicolor L. in China (Zhang & Zhang, 2006). This is the first report of the species as an endophyte on Q. aegilops, Q. infectoria, and in the world. Additionally, the species is reported for the first time in Iraq.
Molecular characterization of Alternaria species
The positive PCR products were sent for sequencing (both forward and reverse primers of single plex PCR and semi-nested PCR) to Macrogen Company in South Korea. DNA sequencing was carried out in both directions, forward and reverse sequences which were edited and assembled, and the consensus sequence was derived for each sample using BioEdite (www.mbio.ncsu.edu). For the determination of Alternaria species, the sequences were subjected to BLAST (https://blast.ncbi.nlm.nih.gov/Blast) searches at NCBI.
All sequences for the ITS1 gene were deposited in Genbank under the accession numbers ON923661, ON923657, ON923663 for A. sorghi, ON923655 for A. consortialis, and ON923670 for A. malorum with 99.30%–100.00% homology with published sequences in GenBank from China, Jordan, Saudi Arabia, Iran, Algeria, and Iraq (Table 2).
Table 2
Details used in the phylogenetic tree for the Alternaria species based on the ITS primer.
All sequences for the LSU gene were deposited in Genbank under the accession numbers OQ160800, OQ160799, OQ160801, OQ152478.1, OQ160817 for A. alternata, A. doliconidium, A. alternata, A angustiovoidea, and A. tenuissima, respectively, with 93.19%–100.00% homology with published sequences in GenBank from South Korea, South Africa, Thailand, The 10 Alternaria nucleotide sequences currently available were subjected to phylogenetic analysis, together with the other published Alternaria species in GenBank (Figure 8, Figure 9), which displayed the findings of this investigation (Table 2, Table 3).
Figure 8
Phylogenetic tree based on the ITS1 gene partial sequencing of Alternariaspp. isolated from Quercus aegilops and Quercus infectoria (leaf, bud) indicated with red arrows. The Neighbor Joining tree was aligned using the Muscle method associated with nucleotide substitution models (K2: Kimura 2 parameter). The tree was made using MEGAX VERSION 10 with a bootstrap value of 1,000 replicates. Numbers on the nodes are bootstrap values.

Figure 9
Phylogenetic tree based on the LSU gene partial sequencing of Alternaria spp. isolated from Quercus aegilops (leaf, bud) indicated with red arrows. The Neighbor Joining tree was aligned using the Muscle method associated with nucleotide substitution models (JC: Jukes–Cantor). The tree was made using MEGAX VERSION 10 with a bootstrap value of 1,000 replicates. Numbers on the nodes are bootstrap values.

Table 3
Details used in the phylogenetic tree for the Alternaria species based on LSU primer.
The phylogenetic analysis based on the ITS gene showed that the Alternaria spp. from Quercus aegilops and Q. infectoria are genetically distinct. Alternaria malorum with accession number ON923670 forms a well-supported clade (100% bootstrap) with other isolates (MZ540328, MF055672, and OP006224), while A. consortialis with accession number ON923655 forms a separate clade with isolate MN557295 with 73% bootstrap. All three isolates of A. sorghi with accession numbers OQ4330657, ON923663, and ON923661 join the other isolates in the same species in a weakly supported clade.
The phylogenetic inquiry using the LSU gene revealed genetic differences between Alternaria spp. from Q. aegilops and Q. infectoria. A. dolicondium with accession number OQ160799 forms a well-supported clade (100% bootstrap) with other isolates, while both isolates of A. alternate with accession numbers OQ160800 and OQ160801 form a separate clade with other A. alternate isolates with 53% bootstrap. Both A. angustiovoidea (OQ 152478) and A. tenuissima (OQ160817) represented an outgroup in the tree constructions.
. Conclusion
The present study represents the first report of endophytic Alternaria species associated with two oak plant species that dominate in the mountainous region of Kurdistan, Iraq. We identified seven endophytic Alternaria species either from Q. aegilops or Q. infectoria based on morphological and molecular characteristics.