. Introduction
Gametophytic apomixis – the parthenogenetic production of offspring from the cell (usually an egg) of an unreduced embryo sac – is a widespread and largely unexplained phenomenon. It is very common in Taraxacum and Hieracium, two large genera of Asteraceae. Both genera include sexually reproducing diploid, strictly self-incompatible, insect-pollinated species and a huge number of polyploid taxa (agamospecies), mainly tri- and tetraploid, which reproduce apomictically (Fehrer et al., 2009; Hörandl, 2018; Kirschner & Štepánek, 1996; Mráz et al., 2019; Pinc et al., 2020; Skalińska, 1970; Tas & van Dijk, 1999; van Baarlen et al., 2000). They share some important similarities: the origin of the apomictic embryo sac (diplospory) and the ability to produce partially fertile pollen grains. Furthermore, they are near-obligatory apomicts producing the embryo parthenogenetically and endosperm autonomously (Bicknell & Koltunow, 2004; Koltunow et al., 2011; Tas & van Dijk, 1999). The presence of sexual diploids and the production of fertile pollen by polyploids facilitated hybridization and the emergence of new apomictic lineages again capable of occasional crossing with diploid plants (de Kovel & de Jong, 2000; Hörandl, 2018; Majeský et al., 2012; Richards, 1970; Shibaike et al., 2002). It increases the evolutionary dynamics of agamic complexes and results in apomictic hybrid swarms. The most interesting feature of the described processes is that the allopolyploids formed in this way can inherit an asexual mode of reproduction from a male parent (via pollen) (Bicknell & Koltunow, 2004; Richards, 1970, 1973).
In the absence of apomixis in diploids, it can be assumed that the trigger for such processes was the emergence of primordial triploid agamospecies, most probably of hybrid origin (Belyayev et al., 2018; Hojsgaard, 2018; Richards, 2003). These plants become donors of reduced and unreduced pollen grains and parents of hybrid offspring with different ploidy levels and variable modes of reproduction (Morita et al., 1990; Tas & van Dijk, 1999). Interestingly, apomixis is transmitted only by the unreduced or not fully reduced pollen grains containing more than one chromosome set (Bicknell & Koltunow, 2004).
While some studies point to its relatively simple pattern of inheritance, obligatory apomixis is almost certainly a complex phenomenon influenced by unlinked gene loci (Hojsgaard, 2020; Niccolo et al., 2023). According to Carman’s (1997) duplicate-gene asynchrony hypothesis, the main cause of embryological anomalies associated with this type of reproduction is the hybridization-derived floral asynchrony triggered by the mismatch of parental sets of developmental genes. Consequently, the appearance of highly specific apomixis-determining genes is not required and some later acquired gene mutations can only improve the seed set and fitness of apomictic lineages. Carman (2001) formulated the essence of his hypothesis as follows: “Apomixis appears to result from competition between genetically-balanced genomes, i.e., nearly complete cassettes of genes”. This approach provides the best explanation for both the transmission of apomixis only by gametes possessing two or more chromosome sets and a seemingly simple pattern of its inheritance. This assumption was supported by experimental crosses between diploid sexuals × triploid apomicts in Taraxacum obtained by Tas and van Dijk (1999). The authors showed the presence of true 2x, 3x and 4x hybrids of different modes of reproduction among progeny and proved that all obtained tetraploids and 1/3 of triploids were capable of apomictic reproduction, whereas diploids were not. This showed that in some cases, the two genomes of a triploid parent may be sufficient to pass apomixis to the offspring, but reliable (100%) inheritance of this reproductive trait is only guaranteed by gametes containing the complete chromosome set of the apomictic parent. Hence, when it is possible for once-formed apomictic plants to crossbreed with diploids (serving as female parents), one can expect the emergence of separate asexually reproducing ancestor-descendant lines of an increasing ploidy level.
Since, so far, no case of fully functional apomixis has been observed in a natural or experimental hybrid of entirely sexual plants, it can be assumed that this is a rare phenomenon requiring a very unique combination of parental genomes. According to Carman (1997), the most favorable conditions for it occurred in the Pleistocene and it was facilitated by extensive changes in ranges of species differing significantly in the timing of the developmental processes. It is not known how often such events took place within a given genus and how many separate apomictic lineages were generated. In the genera highly abundant in apomicts, the correct identification of separate apomictic strains could simplify taxonomic and phylogenetic research.
The genus Hieracium is divided into sections and informal groups of species on the basis of morphological similarities (Stace, 1998). The sections were formed around the so-called main (basic) species, from which all the other species originated as a result of hybridization (Zahn, 1921–1923). According to contemporary authors, the genus comprises a limited number of basic species composed of groups of diploid and polyploid plants characterized by similar morphology and a multitude of intermediate polyploid forms (J. Chrtek et al., 2020; J. Chrtek, Jr. et al., 2009; Fehrer et al., 2009; Mráz et al., 2019). It is assumed that hybridization concerned a small number of diploid species, presumably with quite a wide primary range (Merxmüller, 1975). Currently, the genus Hieracium is dominated by apomictic polyploids (3x–5x), whereas the basal diploid species are sparse and occur mainly in refugial areas of southern Europe (Chrtek, 1997; Fehrer et al., 2009; Ilnicki & Szeląg, 2011; Merxmüller, 1975; Mráz & Zdvořák, 2019; Musiał & Szeląg, 2015; Szeląg, 2010; Szeląg et al., 2007; Szeląg & Ilnicki, 2011; Vladimirov & Szeląg, 2006).
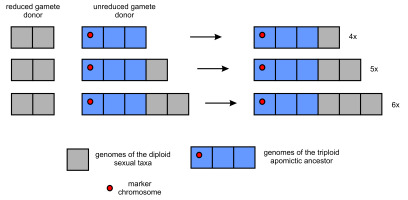
The occurrence of morphologically identifiable basic taxa, which can be considered separate units of evolution (J. Chrtek, Jr. et al., 2009), makes this genus a useful model for studying apomictic complexes. Under the simplicist model discussed above, each separate polyploid series could represent the ancestor-descendant line derived from the primary (most probably triploid) apomictic plant. If any of the genomes of this primordial form contained a detectable molecular or chromosomal marker, it should be presented in all apomictic plants within this series, regardless of their ploidy level (Figure 1). Unfortunately, there is not enough data to support this assumption. Research conducted with the use of molecular cytogenetics methods (fluorescence in situ hybridization - FISH) should provide information in this regard, but it has been scarce and focused on other issues. Only seven diploid and three triploid forms were analyzed with FISH (fluorescence in situ hybridization) using 5S rDNA, 45S rDNA or various satDNA probes, and six - four 3x and two 4x with GISH (genome in situ hybridization) with gDNA probes from selected diploid species (Belyayev et al., 2018; J. Chrtek et al., 2020; Fehrer et al., 2021; Grabowska-Joachimiak et al., 2021; Ilnicki et al., 2010; Mráz et al., 2019; Zagorski et al., 2020).
Figure 1
Probable genome combinations in polyploid descendants of a primary triploid apomictic plant. Each square represents a single genome.
All 10 Hieracium taxa tested in terms of the distribution of the FISH-detectable rDNA segments within the karyotype showed the presence of the characteristic satellite (SAT) chromosomes with two adjacent rDNA clusters (5S and 45S) on the short arm (usually one per genome, hereinafter referred to as the “standard 5S/45S chromosome”) and in addition chromosomes with a terminally located 45S rDNA cluster (1–2 per genome) (Fehrer et al., 2021; Grabowska-Joachimiak et al., 2021; Mráz et al., 2019; Zagorski et al., 2020). However, in the triploid H. telekianum, three chromosomes with differently localized 5S rDNA were observed, of which only one also showed the 45S rDNA (Mráz et al., 2019). According to these authors, the presence of three different chromosomes with 5S rDNA suggests that they belong to three different haplomes (single chromosome sets). Therefore, it seems that more extensive FISH-aimed rDNA analysis may provide simple and easy-to-use chromosomal markers in this genus.
During the initial testing of Hieracium karyotypes in this respect, we observed the atypical chromosome containing 5S rDNA in the karyotype of a plant belonging to the H. sect. Alpina. Because this section can be regarded as one of the basic taxa within the genus Hieracium and includes (apart from the diploid H. alpinum) all polyploid levels (3x–5x) recorded so far in Hieracium s.str., we decided to check if this atypical chromosome would be found in all of them. This would support the assumption concerning the above-mentioned mechanism by which the ploidy level increases within the separate apomictic lineages (Figure 1). For comparison, we also analyzed two triploid species classified in H. sect. Bifida, from which only polyploid forms were known, to check whether they have the same or different rDNA distribution pattern.
. Material and methods
Plant material
The research was conducted on four cytotypes of H. sect. Alpina: H. alpinum 2x, H. alpinum 3x, H. schustleri 4x, and H. chrysostyloides 5x (Table 1). Hieracium sect. Alpina comprises the high mountain species growing in the Alps, Carpathians, Sudetes, Ural, Scandinavia and Greenland (Bräutigam, 1992), its main species is H. alpinum known from diploid and triploid populations (Mráz et al., 2009). In the case of the diploid H. alpinum, plants from two populations that differed with respect to the color of achenes (black vs. brown) were also considered, whereas in the case of H. schustleri plants differing in the color of styles (yellow vs. dark) were analyzed, in order to check whether the specimens with these unusual traits are karyotypically different from the typical ones (with black achenes and yellow styles). Such color variants have not been previously observed in the above-mentioned species. Hieracium alpinum 3x, H. schustleri 4x, H. chrysostyloides 5x occur sympatrically in the alpine zone of the Sudetes. The diploid populations of H. alpinum are presently known only from the refugial areas in the Eastern and Southern Carpathians. Their occurrence, however, in the Western Carpathians and Sudetes before the last Pleistocene glaciation was highly possible.
Table 1
The origin of Hieracium plants used for FISH analysis.
Hieracium sect. Bifida was represented by two triploid taxa: H. bifidum and H. levicaule (Table 1). Hieracium sect. Bifida includes thermophilic grassland plants which occur in lowland regions and in the mountains at lower altitudes. The main species of the section is H. bifidum, widespread in Europe and known from triploid and tetraploid populations (Musiał et al., 2016).
Herbarium specimens of all analyzed forms are deposited in KRAM (National Biodiversity Collection of Recent and Fossil Organisms at W. Szafer Institute of Botany, Polish Academy of Sciences).
All the analyzed plants were obtained from seeds sampled in natural populations and cultivated in a vegetation room of the Department of Plant Breeding, Physiology and Seed Science, University of Agriculture in Krakow. The growing conditions were described by Grabowska-Joachimiak et al. (2021). For each of the studied forms, radicles taken from 10–15 individuals were used to make chromosome slides.
Chromosome preparation and rDNA-FISH
Radicles collected from the cultivated plants were pretreated with a saturated solution of α-bromonaphthalene for 24 h and fixed in a mixture of glacial acetic acid and absolute ethanol (1:3, v/v). Before squashing in 45% acetic acid, root tips were macerated enzymatically (4% pectinase + 2% cellulase in 0.01 M citric buffer, pH 4.6–4.9) at 37 °C for 15 min (H. alpinum, H. chrysostyloides, H. bifidum) or for 20 min (H. levicaule, H. schustleri). The squashes were frozen, air-dried, then stored at −20 °C.
In the FISH procedure, two rDNA probes were used. For visualization of 45S rDNA loci, the probe was based on a 2.3 kb ClaI subclone representing the fragment of a 25S rDNA genic region of Arabidopsis thaliana (Unfried & Gruendler, 1990), labeled by nick translation with tetramethyl-rhodamine-5-dUTP (Roche). The probe for detection of 5S rDNA loci was generated by PCR amplification of a 410 bp fragment of 5S rDNA unit isolated from wheat from the pTa794 clone (Gerlach & Dyer, 1980) and labeled with digoxigenin-11-dUTP (Roche). Amplification was performed using a T-100 Thermal Cycler (Bio-Rad) and a PCR program that consisted of initial denaturation for 1 min at 94 °C, followed by 35 cycles of denaturation for 40 s at 94 °C, annealing for 40 s at 55 °C, an extension for 1 min at 72 °C and a final extension of 5 min at 72 °C. The FISH procedure was performed as described in detail by Wolny and Hasterok (2009). The chromosome preparations, together with the hybridization mixture, were denatured at 70 °C for 5 min and allowed to hybridize for 18 h in a humid chamber at 37 °C. After that, slides were washed in two changes of 10% formamide in 0.1x SSC (saline sodium citrate), for 5 min each, at 42 °C (79% stringency) and subsequently in three changes of 2x SSC for 3 min each, at the same temperature. The digoxigenated probe was immunodetected according to standard protocols by anti-digoxigenin antibodies conjugated with fluorescein isothiocyanate (FITC; Roche). The preparations were counterstained with 4′,6-diamidino-2-phenylindole (DAPI), mounted in Vectashield (Vector Laboratories). 10–20 well-labelled metaphase plates per taxon were analyzed.
FISH images were acquired using a Nikon DS-2MBWc camera and the NIS Elements software attached to a Nikon Eclipse E800 microscope. For image processing, ImageJ 1.46g software (https://imagej.nih.gov/ij) was used. For better visualization of very tiny FISH signals, we used the option ‘find edges’ of the ImageJ program.
. Results
All FISH-detected rDNA signals within Hieracium karyotypes were localized on the short chromosome arms. The 45S rDNA sites were observed at the chromosome ends, and 5S rDNA loci at the pericentromeric position. Three stable (observed in all plants belonging to a given taxon) types of rDNA-bearing chromosomes were observed: (i) chromosomes showing both 5S and 45S rDNA signals, (ii) chromosomes showing only a 45S rDNA signal, and (iii) chromosomes showing only a 5S rDNA signal. They are hereinafter referred to as “standard 5S/45S chromosomes”, “45S chromosomes” and “5S chromosomes”.
Hieracium sect. Alpina
In the karyotype of the diploid H. alpinum (2n = 2x = 18), two standard 5S/45S chromosomes and two 45S chromosomes with a large rDNA segment were observed (Figure 2A–B). Thus, the single standard genome of the H. alpinum group contained two chromosomes with major (well visible in all metaphases) rDNA sequences detectable by FISH procedure: one standard 5S/45S chromosome and one 45S chromosome. Moreover, three plants obtained from brown-colored achenes possessed an extra 45S rDNA segment on the short arm of one chromosome (Figure 2B).
Figure 2
FISH on Hieracium chromosomes with 5S rDNA (red) and 45S rDNA (green) probes, the chromosomes were counterstained with DAPI (blue). (A) Diploid H. alpinum (black achenes); (B) Diploid H. alpinum (brown achenes), note a small extra 45S rDNA segment indicated with arrow; (C) Triploid H. alpinum; (D) Tetraploid H. schustleri (dark styles); (E,F) H. schustleri (yellow styles); image (F) was processed with the use of the option “find edges” of the ImageJ program. Asterisks indicate standard 5S/45S chromosome, arrowheads – marker chromosome with 5S rDNA cluster. Scale bar = 5 µm.
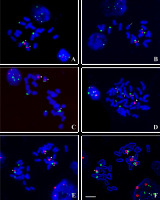
The triploid form of H. alpinum (2n = 3x = 27) had only two standard 5S/45S chromosomes. Next to them, four chromosomes with rDNA sequences were observed: three medium-sized 45S chromosomes and one 5S chromosome (Figure 2C). No extra rDNA signals were observed in the analyzed triploids.
All the tetraploids of H. schustleri plants (2n = 4x = 36) showed three standard 5S/45S chromosomes and one 5S chromosome with a large pericentromeric rDNA cluster. In addition, the progeny of plants with dark styles showed three 45S chromosomes with large, clearly visible 45S rDNA signals (Figure 2D), while that of plants with yellow styles had two 45S chromosomes with a large and one 45S chromosome with a very poor 45S rDNA signal (Figure 2E), which became much more evident after applying the ‘find edges’ option of the ImageJ program (Figure 2F). Thus, in all analyzed tetraploid plants, three 45S chromosomes could be noticed.
The pentaploid H. chrysostyloides (2n = 5x = 45) showed four standard 5S/45S chromosomes, one 5S chromosome, and five 45S chromosomes, all with a large rDNA cluster (Figure 3A). No minor rDNA signals were observed in the karyotype of these plants.
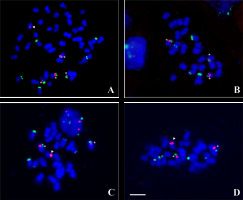
Figure 3
FISH on Hieracium chromosomes with 5S rDNA (red) and 45S rDNA (green) probes, the chromosomes were counterstained with DAPI (blue). (A) Pentaploid H. chrysostyloides; (B) H. bifidum (3x); (C) H. levicaule (3x) from Wyżyna Częstochowska; (D) H. levicaule (3x) from Gorce Mts. Asterisks indicate standard 5S/45S chromosome, arrowheads – marker chromosome with 5S rDNA cluster. Scale bar = 5 µm.
Summing up, all karyotypes of polyploid apomictic plants belonging to the H. sect. Alpina revealed the presence of a single marker 5S chromosome with an rDNA cluster located on the short arm near the centromere. In all these plants, the number of standard 5S/45S chromosomes (i.e., as found in a diploid) is one less than the number of elementary genomes that make up their karyotype (Table 2). The number of the 45S chromosomes was one per genome on three (2x, 3x, 5x) of four ploidy levels. In tetraploid H. schustleri, only three 45S chromosomes were observed. Almost all rDNA signals were large and easily detectable by the FISH procedure used. In some diploid H. alpinum plants, a small additional 45S rDNA signal on the medium-sized chromosome was noticed.
Hieracium sect. Bifida
In the karyotype of the triploid H. bifidum (2n = 3x = 27), three standard 5S/45S chromosomes and four 45S chromosomes were observed (Figure 3B). Thus, there is one standard 5S/45S chromosome per single genome of this plant. No other chromosomes with FISH-detectable rDNA signals were noted.
All the analyzed H. levicaule (2n = 3x = 27) plants showed two standard 5S/45S chromosomes, one 5S chromosome with a large pericentromeric rDNA cluster, and five medium-sized 45S chromosomes. Among specimens from the Wyżyna Częstochowska upland, only two 45S chromosomes showed large rDNA signals (Figure 3C), whereas in plants from the Gorce Mts. (W. Carpathians) - four (Figure 3D).
Two triploids from this section differ in the number of the 45S rDNA-bearing chromosomes per genome (Table 2) and the composition of the karyotype. In H. bifidum, the main species of this section, three standard 5S/45S chromosomes, whereas in the karyotype of H. levicaule, only two chromosomes of this type plus the single 5S chromosome were detected.
. Discussion
In this paper, we showed that all the analyzed polyploid plants belonging to Hieracium sect. Alpina and representing three ploidy levels, have a single marker 5S chromosome in the karyotype. This heterobrachial chromosome possessed a large 5S rDNA cluster on the short arm and was probably a counterpart of the standard 5S/45S chromosome which seems to be a typical component of the Hieracium genome (Fehrer et al., 2021; Grabowska-Joachimiak et al., 2021). This cytogenetic feature confirms the validity of placing the above-mentioned species within one section.
In Taraxacum and Hieracium s.l. (Hieracium s.str. and Pilosella), the increases in the ploidy level seem to be primarily an effect of recursive summing up of genomes of apomictic plants (as donors of unreduced sperm cells) and diploids (as donors of reduced eggs). Since all (or almost all) plants with an elevated ploidy level reproduce apomictically, this trait is most likely the result of inheriting a complete chromosome set containing a unique configuration of developmental genes from an apomictic parent (Carman, 1997). Most probably, the consecutive additions of “sexual” genomes do not change or disturb this developmental path, at least within the observed ranges of ploidy – up to 10x in Taraxacum, 7x in Pilosella, and 5x in Hieracium (Index to Plant Chromosome Numbers: http://legacy.tropicos.org/Project/IPCN). The presence of a single marker 5S chromosome in the analyzed forms of H. sect. Alpina representing successive (3x–5x) ploidy levels confirms this evolutionary mechanism and suggests the 2:1 genome composition in the apomictic ancestor of this group of species. Most probably, two genomes of this plant were derived from the diploid H. alpinum (possessing two standard 5S/45S chromosomes), which is now present only in the Southern and Eastern Carpathians.
The origin of the observed marker 5S chromosome is unclear. Its structure may suggest that it arose from the standard 5S/45S chromosome by losing the terminal part of the short arm containing the 45S rDNA cluster. It is not known, however, whether this occurred at the diploid level or in the primeval triploid apomictic plant. In the first case, this marker chromosome can be passed on to triploid offspring by a reduced or unreduced gamete. If the fused gametes are monospecific, an autotriploid will be produced (Figure 4A). In the majority of the analyzed plants, the rDNA cluster in the 5S chromosome is much bigger than in the standard 5S/45S chromosome, which suggests a strong amplification of 5S rDNA sequences after deletion or during further evolution of the polyploid lineage.
Figure 4
Probable routes of formation of primary triploid apomictic plants with the hemizygous marker chromosome. The scheme takes into account the possibility of the autopolyploid origin of these plants (A), although their hybrid origin and 2:1 genomic composition (B) seem more likely (but see Hojsgaard 2018, and Hojsgaard and Hörandl 2019). Each square represents a single genome.
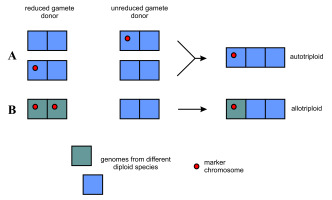
Since it has long been known that most apomictic species are of hybrid origin (Hojsgaard & Hörandl, 2019; Hörandl, 2018; Niccolo et al., 2023, and references therein), it cannot be excluded that the primeval triploid was a hybrid of the sexual forms, one of which had the 5S chromosome in the disomic configuration (Figure 4B). So far, however, no diploid taxon with a pair of 5S chromosomes has been observed (Fehrer et al., 2021; Grabowska-Joachimiak et al., 2021). The observation of a single chromosome similar to the 5S chromosome described here in diploid H. umbellatum from Slovakia (Fehrer et al., 2021) and triploid H. telekianum from Romania (Mráz et al., 2019) may indicate that such a form exists or has existed in Europe. The chromosome in the diploid plant from Slovakia may have arisen through mutation (as suggested by the quoted authors) or hybridization of typical H. umbellatum (possessing two standard 5S/45S chromosomes) with the above-mentioned taxon. The molecular studies by Fehrer et al. (2009) showed that, surprisingly, some diploid Hieracium plants analyzed so far were of hybrid origin. As a result of further crosses involving such plants, progeny of different ploidy levels and different genomic constitution can be created.
Apart from the standard 5S/45S chromosome, the basal chromosome set of different diploid Hieracium species showed one or two other chromosomes with 45S rDNA (45S chromosomes) (Fehrer et al., 2021; Grabowska-Joachimiak et al., 2021). Thus, assuming additivity, the number of these chromosomes in polyploid plants should be at least equal to the number of genomes. There is, however, a clear difference in the number of these chromosomes between the polyploids from Hieracium sect. Alpina and H. sect. Bifida. In Hieracium sect. Alpina their number never exceeds the number of the genomes, whereas in H. sect. Bifida is greater (by one in H. bifidum and by two in H. levicaule) (Table 2). This suggests the different ancestry or different dynamics of 45S rDNA repeats in these two species.
In many polyploid plants, a trend toward reduction in the number of rDNA sites was detected (Garcia et al., 2014; Roa & Guerra, 2012). In the majority of polyploid forms analyzed here, such reduction was not observed. Only in tetraploid H. schustleri, the number of 45S chromosomes was lowered by one (Table 2). A strong reduction in the size of one of the 45S rDNA segments in plants with yellow styles may indicate a trend toward further reduction in the number of 45S rDNA loci in this species.
It has been suggested that the occurrence of gametophytic apomixis in polyploid plants could be related to the presence of a single chromosome containing genes involved in asexual reproduction within a karyotype. Such a hemizygous chromosome was detected after FISH in the apomictic plants belonging to the genus Pilosella and Taraxacum (Kotani et al., 2014; Vašut et al., 2014, and references therein). In Hieracium, however, there is no relationship between the apomixis and the occurrence of the hemizygous marker 5S chromosome described here because part of the apomictic plants analyzed with rDNA probes so far did not have it in their chromosome complements. Also, the two analyzed apomictic taxa from Hieracium sect. Bifida (H. bifidum and H. levicaule) differ in this respect.
Apomictic plants can also produce not fully reduced and even fully reduced (1x) pollen grains, which can lead to hybrid offspring of non-elevated ploidy level and diploids, both showing varying capacity for efficient reproduction (Mártonfiová et al., 2007; Mráz et al., 2019; Tas & van Dijk, 1999; van Dijk et al., 1999). It seems that among the forms thus created, only polyploids which maintained the ability to reproduce apomictically and some sexually reproducing “secondary” diploids are able to survive in nature. All these processes lead to the emergence of a muddle of hybrid taxa of different ploidy levels and a hardly recognizable origin. Attempts to systematize such complexes have been going on for a long time (Mráz et al., 2019, and references therein). One way would be the identification of polyploid series/lines, each of which had a separate apomictic ancestor (most probably triploid), whose unique set of genomes had been preserved without major changes. It is likely that some of these lines may have already been distinguished on the basis of morphological criteria as the so-called basic species or higher-level taxa (e.g., sections). The detected karyotypic differences between triploid H. bifidum and H. levicaule suggest that H. sect. Bifida, unlike the analyzed group of species belonging to H. sect. Alpina is a less homogenous group of species, most probably composed of polyploid forms of different origin. This issue requires clarification in the future.


