Introduction
Heavy metal contamination in soils from cadmium (Cd) is an increasingly urgent problem throughout the world. Cadmium contamination alters the photosynthetic rate and respiration of plants, thereby influencing their profitability and quality (Clemens et al., 2013). Reduction in photosynthetic pigments before CO2 fixation was observed in tomato plants exposed to Cd2+, and this was considered to be the main reason for the reduction in photosynthesis (Baszyński et al., 1980). As one of the most dangerous environmental pollutants, Cd is highly toxic to plants even at low concentrations (Dias et al., 2013; Lin & Aarts, 2012), and leads to a series of stress symptoms including chlorosis, necrotic lesions, wilting, and disturbances in mineral nutrition and carbohydrate metabolism (Rani et al., 2014). After Cd2+ (20 mM) of treatment, wilting occurred in Cajanus cajan and the chlorophyll content decreased by 70%, transpiration rate decreased by 60%, and photosynthesis efficiency decreased by 76% (Sheoran et al., 1990). Previous studies have revealed that high Cd concentrations could denature proteins, leading to cellular damage in higher plants (Lin & Aarts, 2012). Additionally, exposure to 10 mg/L Cd reportedly decreases the total chlorophyll and soluble protein contents of Oenanthe javanica (J. Zhang et al., 2015). Recent molecular studies have shown that when a plant is under Cd stress, the absorption and assimilation of sulfur in the plant will also be affected, thereby affecting the synthesis of sulfur-containing amino acids, protein, and glutathione (GSH) in the plant (Nocito et al., 2006). Some research results show that Cd affects the absorption and accumulation of sulfur by inhibiting the absorption rate and enzyme activity of nitrate (Ferretti et al., 1994). These findings indicate that Cd can damage plants in various ways.
Many plants have evolved multiple effective response mechanisms involving Cd tolerance and detoxification that minimize cellular damage due to heavy metal stress (Gallego et al., 2012; Hall, 2002). These mechanisms include the binding of Cd to the cell wall and the chelation of Cd by some ligands (Clemens, 2001; Gill et al., 2013). However, the cellular activities underlying responses to heavy metal stress vary among plant species. In Phaseolus vulgaris, most of the Cd in the leaves and roots is bound to the cell wall, thereby preventing Cd from damaging the protoplasts (Ni & Wei, 2003). Phytochelatins (PCs) are the main ligands in plants that bind Cd. Accordingly, the PC content determines the Cd tolerance of plants to some extent (Ha et al., 1999). When these detoxification strategies are inadequate, antioxidant metabolism becomes an important mechanism mediating plant response to heavy metals. Superoxide dismutase, catalase, and GSH are antioxidants that are particularly important in plants. Being a central molecule of both the antioxidant defense system and the glyoxalase system, GSH is involved in both direct and indirect control of reactive oxygen species (ROS) and methylglyoxal (MG) and their reaction products in plant cells, thus protecting the plant from heavy metal-induced oxidative damage (Hossain et al., 2012). Studies have shown that after Cd treatment, an increase in the total amount of GSH in plants is observed. When Camellia sinensis responded to Cd stress, it was found that the transcription level of GSH biosynthesis and regeneration genes increased within a certain range as the degree of Cd stress increased (Mohanpuria et al., 2008). The ascorbate–reduced glutathione (AsA–GSH) cycle enables plant cells to eliminate H2O2. The enzymes catalyzing the AsA–GSH cycle in plants are distributed in the chloroplast, cytoplasm, and mitochondria (Drążkiewicz et al., 2003). Many transporters and transcription factors, including MYB, Nramp, bHLH, ZIP, HMA, ABCC, and WRKY, are reportedly involved in Cd detoxification and response in plants (Wang, 2016). In Arabidopsis thaliana, an ABC transporter, PDR8, is a Cd extrusion pump that can inhibit the cytosolic accumulation of Cd (Kim et al., 2007). The overexpression of PDR8 results in increased plant tolerance of Cd and the decreased accumulation of intracellular Cd. Earlier studies confirmed that WRKY transcription factors are important for the responses of higher plants to various abiotic stresses. Additionally, WRKY13 is crucial for plant Cd stress responses (Sheng et al., 2018). Moreover, some key genes have been proven to contribute to responses to heavy metal stress, including OsHMA2, OsNRAMP5, PCS1, ATM3, and ZNT1 (Lee et al., 2003; Pence et al., 2000; Sasaki et al., 2012).
Potentilla sericea is grown in gardens or as ground cover partly because it produces beautiful flowers. Earlier investigations confirmed that P. sericea could grow normally at a Cd concentration of 1 mg/kg (Wu et al., 2016). However, to optimize the use of P. sericea for repairing soil contaminated with heavy metals, the mechanism underlying P. sericea responses to Cd stress must be more thoroughly characterized. Transcriptomics techniques have recently become more widely used for studying plant responses to various abiotic stresses. These types of investigations can clarify plant gene expression and enable the discovery of new functional genes. For example, a transcriptomics analysis was applied to elucidate the mechanism responsible for the Brassica napus response to Cd stress (Gao, 2013). In a recent transcriptomic study of Sedum alfredii exposed to Cd stress, many key genes related to Cd hyperaccumulation were identified (Sheng et al., 2018). Therefore, in the current study, the Cd response of P. sericea was evaluated based on a physiological examination and the application of digital gene expression technology to compare the gene expression between P. sericea roots treated with 90 μmol/L Cd2+ for 7 days and roots cultured under normal conditions. Our data revealed that P. sericea responds to Cd stress. The results of this study may be useful for identifying and characterizing the genes and pathways mediating the Cd response of P. sericea.
Material and Methods
Material and Treatments
The test materials were planted in the nursery test station of the College of Landscape Architecture of Northeast Forestry University (China) and managed conventionally. The P. sericea seedlings, kept free of disease and pests for half a year, were harvested in October 2018. The samples were thoroughly washed with sterile water and dried, after which two uniformly growing healthy P. sericea seedlings were added to Erlenmeyer flasks containing half-strength Hoagland’s nutrient solution to induce further growth. The nutrient solution was refreshed every 3 days until the plants exhibited strong growth. Previous studies have proved that the aboveground P. sericea plant parts are severely affected by a 25-day treatment with 20 mg/L Cd2+ solution, whereas an approximately 1-week treatment only results in the yellowing of the leaf edges (J. Zhang et al., 2015). Thus, this time-point was selected for analyzing the effects of 0 (control) and 90 μmol/L Cd2+ (i.e., Cd0 and Cd90 treatments, respectively) on the P. sericea transcriptome and physiological characteristics. Cd can only exist in the soil in the form of +2 valent simple ions or simple coordination ions. Therefore, +2 valent Cd was used in the experiment. Cd is added in the form of CdCl2·2.5H2O (J. Zhang et al., 2015). At 7 days after initiating the treatments, the roots and leaves healthy P. sericea seedlings grown in the same position were collected in the morning for subsequent experiments. The treatments were completed with three biological replicates.
Photosynthesis Rate Determination
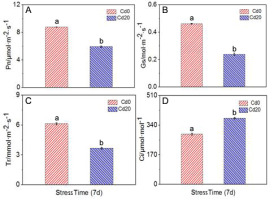
A portable photosynthesis system that measures CO2 uptake (LCi, Leaf Chamber Analysis System; ADC Bio Scientific, Hoddesdon, UK) equipped with a square (6.25 cm2) chamber was used for determinations of net photosynthetic rate (Pn, µmol m−2 s−1), transpiration rate (Tr, mmol m−2 s−1), stomatal conductance to water vapor (Gs, mol m−2 s−1), and intercellular CO2 concentration (Ci, µmol mol−1) of P. sericea.
Total RNA Isolation and Transcriptome Sequencing
Total RNA was extracted from the Cd0 and Cd90 roots with the TRIzol reagent. The quality and concentration of the RNA were assessed with the NanoDrop 2000 spectrophotometer (Thermo Scientific, USA) and by agarose gel electrophoresis. The extracted RNA was used as the template to synthesize single-stranded cDNA with SuperScript Reverse III Transcriptase (Invitrogen, USA). The resulting cDNA library was analyzed with the Illumina HiSeq 2500 high-throughput platform, and sequences were assembled with Trinity to construct a sequencing library. The sequencing and assembly of the library were completed by Beijing Baimaike Biotechnology.
Gene Functional Annotation and Expression Analysis
Unigenes were functionally annotated according to BLAST-mediated (Altschul et al., 1997) comparisons with the sequences in the RefSeq nonredundant proteins (Nr) (Li et al., 2002), COG (Tatusov et al., 2000), Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa et al., 2004), and Swiss-Prot (Apweiler et al., 2004) databases. Unigenes were annotated with gene ontology (GO) terms based on the GO terms assigned to the corresponding Swiss-Prot sequences (Bolger et al., 2014). The gene expression levels, which were represented by the fragments per kilobase of transcript per million mapped reads (FPKM), were analyzed with the Bowtie2 and eXpress programs (Langmead & Salzberg, 2012).
Differentially Expressed Gene (DEG) Functional Annotation and Analysis
Differentially expressed genes were detected based on the following criteria: false-discovery rate < 0.005 and an absolute log2 fold change ≥ 2. All DEGs in the P. sericea transcriptome were compared with sequences in the GO, COG, and KEGG databases to identify significantly enriched functions and pathways (p < 0.05) (Wixon & Kell, 2000).
Validation by Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Six DEGs were randomly selected form metal ion transporters for a qRT-PCR assay. On the basis of alignments with sequences in the NCBI database, genes associated with metal ion transporters were identified in the P. sericea transcriptome. Appropriate primers for these genes were designed with Vector NTI Advance 11.0. The qRT-PCR assay was completed with the RealPlex2 real-time fluorescent PCR system (Eppendorf, Germany) and SYBR Premix Ex Taq II (Perfect Real Time, Takara, Japan). The PCR program was as follows: 95 °C for 30 s; 40 cycles of 95 °C for 5 s, 60 °C for 15 s, and 72 °C for 30 s.
Results
Photosynthesis Under Cd Stress Conditions
The net photosynthetic rate (Pn) in the Cd treatments was lower than in control treatments by an average of 32.4% over the study. Transpiration rate (Tr) was higher in the control treatment by 40% compared with the Cd treatment. The intercellular concentration of CO2 (Ci) was higher in Cd treatments by an average of 31.9%. Stomatal conductance of water vapor (Gs) was lower in the Cd treatments compared with the control by an average of 47.8% (Figure 1) (p < 0.05). Although a high concentration of Cd stress reduces the photosynthesis rate, the treatment group still survived after 1 week, although the leaves turned yellow (Figure 2).
Root Transcriptome Under Cd Stress Conditions
The filtering of the raw sequencing data resulted in 78.06 Gb clean sequencing data for the three biological replicates of the Cd0 and Cd90 samples. A total of 94,009 transcripts and 53,225 unigenes were obtained after the sequences were assembled. The N50 values for the transcripts and unigenes were 2,082 and 1,946, respectively, which were indicative of a relatively high sequence assembly integrity. The functional annotation of unigenes based on comparisons with sequences in the Nr, Swiss-Prot, KEGG, COG, and GO databases resulted in 43,991 annotated unigenes.
Analysis of DEGs
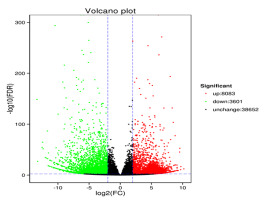
On the basis of an adjusted p < 0.05, 11,684 genes were identified as significantly differentially expressed between the control (Cd0) and treated (Cd90) P. sericea plants. Among these DEGs, 8,083 were upregulated and 3,601 downregulated. A volcano plot of the DEGs is presented in Figure 3.
Figure 3
Volcano plot of differentially expressed genes between control (Cd0) and treated (Cd90). Red and green dots indicate the upregulated and downregulated genes, respectively. Black dots indicate the genes that were showed no significant differences in expression between control and Cd-treated samples.
GO Enrichment Analysis of DEGs
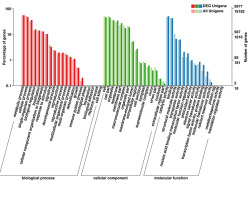
A total of 5,877 DEGs were functionally annotated based on the GO database. These annotated DEGs comprised 3,813 upregulated and 2,064 downregulated genes, which were divided into the three main GO categories as follows: 4,426 annotated DEGs in the biological process category; 4,626 DEGs in the molecular function category; and 4,014 DEGs in the cell component category (Figure 4). The GO functional enrichment analysis indicated that many cellular processes (e.g., cells and cellular parts) were significantly enriched among the DEGs. These processes can lead to changes in cell wall components that affect their ability to bind to heavy metals. Many biological processes related to plant signal transduction, such as the metabolic process of chitin (GO:0006030) and glutamate receptor signaling pathway (GO:0007215) and the related genes are upregulated, implying plant responses to heavy metal stress requires the activation of a signal transduction pathway and the related enzymes.
Figure 4
Differentially expressed genes (DEGs) between sample groups and gene ontology (GO) functional annotations of all genes. GO classification according to biological process, cellular component, and molecular function categories of all annotated 18,182 unigenes and 5,877 differentially expressed unigenes. Subcategories are indicated on the x axis. The right y axis represents the number of genes in each category, and the left y axis represents the percentage of genes in each GO category that belong to a specific subcategory.
COG Annotation Analysis of DEGs
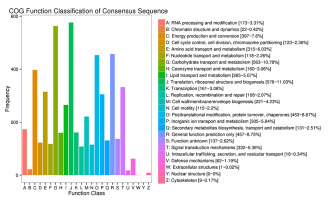
The annotation of the DEGs in the P. sericea transcriptome based on the COG database is summarized in Figure 5. The DEGs were classified into 25 categories, with genes associated with the translation and ribosomal structure and biogenesis accounting for the highest proportion, followed by genes associated with carbohydrate transport and metabolism. These genes most likely include those associated with Cd2+ response and detoxification. In contrast, relatively few genes were related to extracellular and nuclear structures.
Figure 5
Differential expression gene COG annotation results. All putative proteins were aligned to the COG database and functionally classified into one or more of 25 molecular families. The capital letters on the x axis correspond to the COG categories listed to the right of the histogram, and the y axis indicates the numbers of differentially expressed genes assigned to the corresponding COG category.
KEGG Enrichment Analysis of DEGs
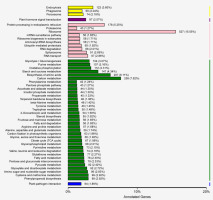
An analysis of the KEGG pathways enriched among the DEGs revealed 127 metabolic pathways. A total of 527 DEGs were involved in ribosome metabolism (ko03010). Other enriched KEGG pathways included carbon metabolism (ko01200, 258 DEGs), amino acid biological synthesis (ko01230, 207 DEGs), protein processing in the endoplasmic reticulum (ko04141, 178 DEGs), and starch and sucrose metabolism (ko00500, 147 DEGs) (Figure 6). Additionally, the P. sericea transcriptome included DEGs involved in secondary metabolism-related pathways (i.e., phenylpropanoid biosynthesis and flavonoid biosynthesis). By comparing the transcriptome data of the Cd-stressed and control P. sericea plants, we identified 108 DEGs related to glutathione metabolism (ko00480, 77 DEGs), sulfur metabolism (ko00920, 31 DEGs), all of which are likely to contribute to Cd stress responses and cellular detoxification processes (Wixon & Kell, 2000). The expression levels of genes encoding enzymes in the glutathione metabolic pathways, such as glutathione peroxidase and glutathione S-transferase (GST), are downregulated during Cd stress. Additionally, genes related to GST (c399387.graph_c0) and glutathione synthetase (GS; c413509.graph_c0) were differentially expressed in the Cd0 and Cd90 samples. The expression of related genes may be part of a P. sericea strategy to protect against Cd stress. The gene encoding ATP-sulfurylase (ATP-S) (c410929.graph_c0), which enables SO42− to enter the sulfate assimilation pathway, was also differentially expressed between Cd0 and Cd90 samples. The expression levels of the genes encoding the other enzymes involved in subsequent steps, including APS reductase (APR) (c397270.graph_c0), sulfite reductase (c412984.graph_c1), and O-acetylserine (thiol) lyase, were revealed as upregulated in the Cd0 vs. Cd90 comparison. These enzymes play a key role in sulfate metabolism.
Figure 6
Distribution of KEGG pathway involved in Potentilla sericea. The y axis represents the name of the KEGG pathway and the number and proportion of differential genes under the pathway. The yellow bars correspond to KEGG pathway involved in cellular processes; purple bars correspond to KEGG pathway involved in environmental information processing; pink bars correspond to KEGG pathway involved in genetic information processing; green bars correspond to KEGG pathway involved in metabolism; blue bars correspond to KEGG pathway involved in organismal systems.
Metal Ion Transporter Genes Responsive to Abiotic Stimuli
According to the results of a BLASTP functional annotation comparison, 21 differentially expressed transporter genes related to metals (e.g., Ca, Zn, Cu, Fe, Na, K, and Cd) were screened from the transcriptomes of the control and Cd-treated P. sericea plants. Of these 21 genes, 13 exhibited upregulated expression, and eight exhibited downregulated expression (Table 1).
Table 1
Cd-stress induced metal transporter genes in root of Potentilla sericea.
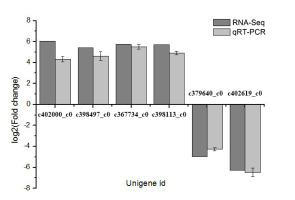
Validation of DEGs by qRT-PCR
To verify the transcriptome data, six DEGs were randomly selected for a qRT-PCR analysis. The results of the qRT-PCR assay, which complied with the minimum information for the publication of quantitative real-time PCR experiments (MIQE) guidelines, were consistent with the P. sericea transcriptome data, with similar expression trends detected (Figure 7). Therefore, the transcriptome analysis was confirmed as reliable.
Discussion
In this study, we identified numerous DEGs associated with a complex network of genes controlling Cd stress responses. Additionally, our data provide insights into the mechanism underlying the P. sericea response to Cd stress and may represent a valuable genetic resource useful for breeders interested in producing transgenic P. sericea resistant to heavy metals.
In plants, Cd induces oxidative stress and inhibits photosynthetic and growth responses of plants. Cadmium toxicity negatively influences photosynthesis by reducing stomatal conductance and stomatal frequency (Mobin & Khan, 2007). In this study, a Cd2+ treatment reduced photosynthesis in P. sericea and resulted in chlorotic leaves. In the transcriptome data, a total of 183 DEGs were related to chloroplasts (GO:0009507) based on the GO database.
Analysis of the KEGG pathway enriched in DEG revealed that 25 genes were involved in photosynthesis (ko00195). Among them, 24 genes related to photosynthesis were downregulated. This indicated that the growth of the plant was inhibited by Cd stress, and the plant leaves may have turned yellow due to Cd poisoning. Other studies confirmed that cell walls are important for plant detoxification (Cosio et al., 2005; Vollenweider et al., 2006). An exposure to Cd2+ stress may inhibit the binding of heavy metals to cellular protoplasts because of changes to certain cell wall components that enhance their ability to bind heavy metals. The GO functional enrichment analysis indicated that 191 of the identified DEGs are related to the cell wall (GO:0005618). A number of signaling pathways lead to differential gene regulation. Previous studies have also reported the activation of the MAPK signaling by Cd (Yeh et al., 2006). The MAPK signaling cascade modulates homeostasis of ROS, stress adaptation, and tolerance in plants. In the present study, the MAPK signaling cascade activated a wide range of kinases and transcription factors such as WRKY 33, ERF1, which can further induce a wide array of stress and growth responses. Modulation of gene expression of signaling pathways components indicates their involvement in managing Cd stress in the present study.
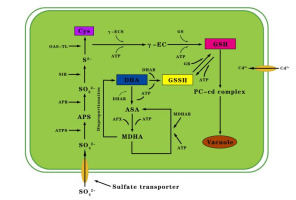
The KEGG pathways with significant enrichment of DEGs are mainly ribosome metabolism (ko03010) and carbon metabolism (ko01200). Ribosomes are organelles for protein synthesis, and their only function is to efficiently and accurately synthesize peptide chains from amino acids according to the instructions of mRNA (Wang, 2016). The expression of related genes under Cd stress may affect the process of translation of mRNA into protein. As the most basic and most important physiological and biochemical reaction of plants, carbon metabolism can provide the materials and energy needed for plant growth and metabolism, and enhance the ability of plants to adapt to stress (Sun et al., 2015). Additionally, Cd stress significantly affected the expression of genes involved in phenylpropane biosynthesis pathways. Phenylpropanoids contribute to all aspects of plant responses towards biotic and abiotic stimuli and contribute to the synthesis of phenylpropanoid-based polymers (Vogt, 2010). Phenylpropanoid-based polymers, like lignin, can enhance the toughness of plant cell walls, thereby enhancing plant defense and detoxification of heavy metals (Z. Zhang et al., 2011). The latest reports have highlighted the potential role of phenylpropanoid pathway metabolites, especially flavonoids, as effective antioxidants. The activity of phenylalanine ammonia-lyase (PAL; c415593_c0) and chalcone synthase (CHS; c412475.graph_c0), key enzymes in the flavonoid synthesis, is affected by various environmental stimuli, including heavy metals. Stimulation of CHS activity has already been observed in plants exposed to copper and Cd (Izbiańska et al., 2014). Previous studies have proved that AtHMA2 and OsHMA2 in A. thaliana and rice, respectively, enhance the ability of plants under Cd stress conditions to absorb and transport Cd (Satoh-Nagasawa et al., 2011; Takahashi et al., 2012; Wong & Cobbett, 2009). The P. sericea homologs of AtHMA2 and OsHMA2 (c385727.graph_c2 and c411337.graph_c0, respectively) were detected in the transcriptome, suggesting c385727.graph_c2 and c411337.graph_c0 are important for the uptake and transport of Cd in P. sericea. The PaHMA2 gene (c385727.graph_c2 and c411337.graph_c0) should be functionally characterized in future studies. The excessive accumulation of metal elements in plants induces oxidative stress, and antioxidants are important for plant responses and Cd tolerance (Jin et al., 1998). Previous research has confirmed that the absorption of sulfur influenced the response to Cd to some extent (Nocito et al., 2006). The uptake of sulfur by plants is important for the production of sulfur-containing amino acids, proteins, cysteine (Cys), and GSH (Spadaro et al., 2010; Rausch & Wachter, 2005). The GSH content of Barbarea orthoceras leaves reportedly increased after the application of exogenous sulfur to Cd-treated plants (Anjum et al., 2008). Studies have proven that low Cd concentrations in Zea mays seedlings increase the ATP-sulfurylase (ATP-S) and adenosine 5-phosphosulfate reductase (AR) contents and activities, with AR catalyzing GSH biosynthesis (Gill & Tuteja, 2011). In an earlier investigation of pea plants, AR and GSH synthetase activities were induced in response to Cd (Adrian et al., 1990). In the current study, the detection of upregulated DEGs related to ATP-S and AR suggested that GSH synthesis increases to enhance the resistance of plants to Cd stress. Specifically, GSH, which is a reducing agent, is oxidized by glutathione reductase (GR) to glutathione disulfide (GSSG) (Adrian et al., 1990). Mediated by enzymes, the interconversion between GSH and GSSG can maintain the dynamic balance of GSH in plants, thereby helping GSH to function as an antioxidant (Rao & Reddy, 2008). Heavy metal stress can disrupt the balance between the production and detoxification of ROS, which alters the normal physiological functions of plants (Bagheri et al., 2017). The clearance of ROS by glutathione is mainly accomplished via three pathways, the most important of which is the ascorbate–glutathione cycle (ASA–GSH). Ascorbate peroxidase (APX) and monodehydroascorbate reductase (MDHAR) are two crucial enzymes in the ASA–GSH cycle. Ascorbate is also an important reducing agent in plants. Furthermore, GSH and dehydroascorbic acid (DHA) are substrates in a reaction catalyzed by dehydroascorbate reductase (DHAR) to form GSSG and ASA. The ASA is oxidized to monodehydroascorbate (MDHA) via a reaction catalyzed by APX, and MDHA is converted to ASA by MDHAR. The important antioxidative effect of GSH occurs through the ASA–GSH cycle (Gill & Tuteja, 2011). The biosynthesis of PC synthase (PCS) is tightly regulated through sulfur metabolism. In plants, both Cys and GSH are involved in PCS synthesis and metal chelation (Figure 8). The application of exogenous GSH leads to differences in GSH and PCS levels (Satoh-Nagasawa et al., 2011). Genes related to sulfur metabolism and glutathione metabolism were searched for in the P. sericea transcriptome is significant for heavy metal stress (Table 2).
Table 2
Cd-induced stress-related genes in root of Potentilla sericea.
Cadmium, a nonessential metal element, enters plant cells via pathways regulating the absorption and transport of other elements. The expression levels of the transporter genes screened in the P. sericea transcriptome were significantly upregulated or downregulated under Cd stress conditions. These transporter genes are likely to play an important role in the absorption and detoxification of heavy metals (e.g., Cd) in P. sericea plants.
Conclusion
Transcriptome sequencing generated comprehensive data regarding DEGs that were reflected in the P. sericea processes induced by Cd stress. The results described herein may enrich the available P. sericea genomic information and serve as a valuable resource for future molecular biology research and the breeding of transgenic P. sericea with greater resistance to heavy metal stress.
Handling Editor
Ewa Sobieszczuk-Nowicka; Adam Mickiewicz University in Poznań, Poland; https://orcid.org/0000-0002-6603-0582